7XQB
 
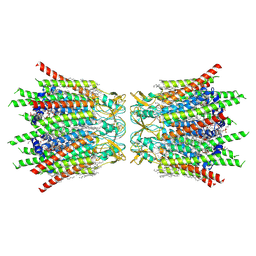 | | Structure of connexin43/Cx43/GJA1 gap junction intercellular channel in POPE/CHS nanodiscs at pH ~8.0 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Gap junction alpha-1 protein, PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Lee, H.J, Cha, H.J, Jeong, H, Lee, S.N, Lee, C.W, Woo, J.S. | | Deposit date: | 2022-05-07 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM.
Nat Commun, 14, 2023
|
|
7XQF
 
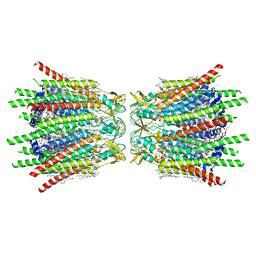 | | Structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel in POPE/CHS nanodiscs | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Gap junction alpha-1 protein, PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Lee, H.J, Cha, H.J, Jeong, H, Lee, S.N, Lee, C.W, Woo, J.S. | | Deposit date: | 2022-05-07 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM.
Nat Commun, 14, 2023
|
|
7XQD
 
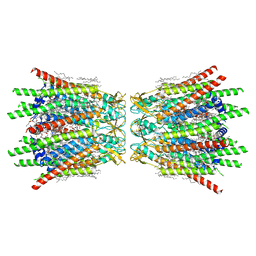 | | Structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel in POPE/CHS nanodiscs (C1 symmetry) | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Gap junction alpha-1 protein, PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Lee, H.J, Cha, H.J, Jeong, H, Lee, S.N, Lee, C.W, Woo, J.S. | | Deposit date: | 2022-05-07 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM.
Nat Commun, 14, 2023
|
|
7XKI
 
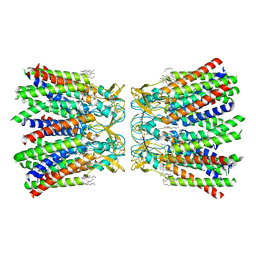 | | Human Cx36/GJD2 (N-terminal deletion BRIL-fused mutant) gap junction channel in soybean lipids (D6 symmetry) | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction delta-2 protein,Soluble cytochrome b562 | | Authors: | Cho, H.J, Lee, S.N, Jeong, H, Ryu, B, Lee, H.J, Woo, J.S, Lee, H.H. | | Deposit date: | 2022-04-19 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of human Cx36/GJD2 neuronal gap junction channel.
Nat Commun, 14, 2023
|
|
7XNV
 
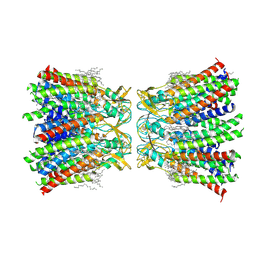 | | Structurally hetero-junctional human Cx36/GJD2 gap junction channel in soybean lipids (C6 symmetry) | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction delta-2 protein | | Authors: | Lee, S.N, Cho, H.J, Jeong, H, Ryu, B, Lee, H.J, Lee, H.H, Woo, J.S. | | Deposit date: | 2022-04-29 | | Release date: | 2023-03-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of human Cx36/GJD2 neuronal gap junction channel.
Nat Commun, 14, 2023
|
|
7XNH
 
 | | Human Cx36/GJD2 gap junction channel with pore-lining N-terminal helices in soybean lipids | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction delta-2 protein | | Authors: | Lee, S.N, Cho, H.J, Jeong, H, Ryu, B, Lee, H.J, Lee, H.H, Woo, J.S. | | Deposit date: | 2022-04-28 | | Release date: | 2023-03-22 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of human Cx36/GJD2 neuronal gap junction channel.
Nat Commun, 14, 2023
|
|
7XL8
 
 | | Human Cx36/GJD2 (N-terminal deletion mutant) gap junction channel in soybean lipids (D6 symmetry) | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction delta-2 protein | | Authors: | Lee, S.N, Cho, H.J, Jeong, H, Ryu, B, Lee, H.J, Lee, H.H, Woo, J.S. | | Deposit date: | 2022-04-21 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of human Cx36/GJD2 neuronal gap junction channel.
Nat Commun, 14, 2023
|
|
7XKT
 
 | | Human Cx36/GJD2 (BRIL-fused mutant) gap junction channel in detergents at 2.2 Angstroms resolution | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Cho, H.J, Lee, S.N, Jeong, H, Ryu, B, Lee, H.J, Woo, J.S, Lee, H.H. | | Deposit date: | 2022-04-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Cryo-EM structures of human Cx36/GJD2 neuronal gap junction channel.
Nat Commun, 14, 2023
|
|
7XKK
 
 | | Human Cx36/GJD2 gap junction channel in detergents | | Descriptor: | Gap junction delta-2 protein | | Authors: | Lee, S.N, Cho, H.J, Jeong, H, Ryu, B, Lee, H.J, Lee, H.H, Woo, J.S. | | Deposit date: | 2022-04-19 | | Release date: | 2023-04-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of human Cx36/GJD2 neuronal gap junction channel.
Nat Commun, 14, 2023
|
|
8HKP
 
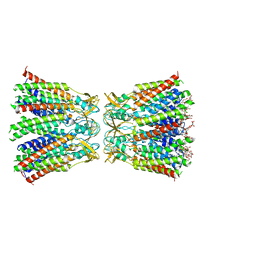 | | Structurally hetero-junctional human Cx36/GJD2 gap junction channel in detergents (C6 symmetry) | | Descriptor: | Gap junction delta-2 protein, Lauryl Maltose Neopentyl Glycol | | Authors: | Lee, S.N, Cho, H.J, Jeong, H, Ryu, B, Lee, H.J, Lee, H.H, Woo, J.S. | | Deposit date: | 2022-11-27 | | Release date: | 2023-03-22 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of human Cx36/GJD2 neuronal gap junction channel.
Nat Commun, 14, 2023
|
|
7SC5
 
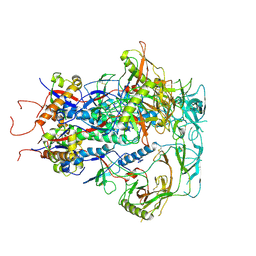 | | Cytoplasmic tail deleted HIV Env trimer in nanodisc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Yang, S, Walz, T. | | Deposit date: | 2021-09-27 | | Release date: | 2022-11-09 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Dynamic HIV-1 spike motion creates vulnerability for its membrane-bound tripod to antibody attack.
Nat Commun, 13, 2022
|
|
7SD3
 
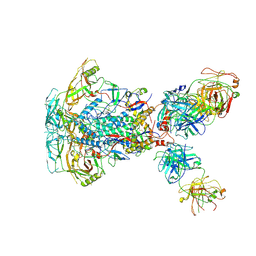 | | Cytoplasmic tail deleted HIV-1 Env bound with three 4E10 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4E10 Fab heavy chain, ... | | Authors: | Yang, S, Walz, T. | | Deposit date: | 2021-09-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Dynamic HIV-1 spike motion creates vulnerability for its membrane-bound tripod to antibody attack.
Nat Commun, 13, 2022
|
|
3H3P
 
 | | Crystal structure of HIV epitope-scaffold 4E10 Fv complex | | Descriptor: | 4E10_S0_1TJLC_004_N, CALCIUM ION, Fv 4E10 heavy chain, ... | | Authors: | Holmes, M.A. | | Deposit date: | 2009-04-16 | | Release date: | 2010-01-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Interactions between lipids and human anti-HIV antibody 4E10 can be reduced without ablating neutralizing activity
J.Virol., 84, 2010
|
|
3VRN
 
 | | Crystal structure of the tyrosine kinase binding domain of Cbl-c | | Descriptor: | CALCIUM ION, Signal transduction protein CBL-C | | Authors: | Takeshita, K, Tezuka, T, Isozaki, Y, Yamashita, E, Suzuki, M, Yamanashi, Y, Yamamoto, T, Nakagawa, A. | | Deposit date: | 2012-04-13 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural flexibility regulates phosphopeptide-binding activity of the tyrosine kinase binding domain of Cbl-c
J.Biochem., 152, 2012
|
|
3VRQ
 
 | | Crystal structure of the tyrosine kinase binding domain of Cbl-c (PL mutant) | | Descriptor: | CALCIUM ION, Signal transduction protein CBL-C | | Authors: | Takeshita, K, Tezuka, T, Isozaki, Y, Yamashita, E, Suzuki, M, Yamanashi, Y, Yamamoto, T, Nakagawa, A. | | Deposit date: | 2012-04-13 | | Release date: | 2013-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural flexibility regulates phosphopeptide-binding activity of the tyrosine kinase binding domain of Cbl-c.
J.Biochem., 152, 2012
|
|
3VRO
 
 | | Crystal structure of the tyrosine kinase binding domain of Cbl-c in complex with phospho-Src peptide | | Descriptor: | CALCIUM ION, Proto-oncogene tyrosine-protein kinase Src, Signal transduction protein CBL-C | | Authors: | Takeshita, K, Tezuka, T, Isozaki, Y, Yamashita, E, Suzuki, M, Yamanashi, Y, Yamamoto, T, Nakagawa, A. | | Deposit date: | 2012-04-13 | | Release date: | 2013-03-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural flexibility regulates phosphopeptide-binding activity of the tyrosine kinase binding domain of Cbl-c.
J.Biochem., 152, 2012
|
|
3VRR
 
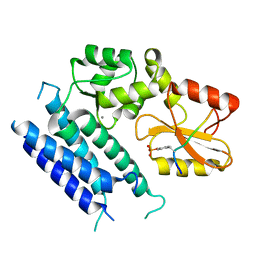 | | Crystal structure of the tyrosine kinase binding domain of Cbl-c (PL mutant) in complex with phospho-EGFR peptide | | Descriptor: | CALCIUM ION, Epidermal growth factor receptor, Signal transduction protein CBL-C | | Authors: | Takeshita, K, Tezuka, T, Isozaki, Y, Yamashita, E, Suzuki, M, Yamanashi, Y, Yamamoto, T, Nakagawa, A. | | Deposit date: | 2012-04-13 | | Release date: | 2013-03-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural flexibility regulates phosphopeptide-binding activity of the tyrosine kinase binding domain of Cbl-c.
J.Biochem., 152, 2012
|
|
2LWK
 
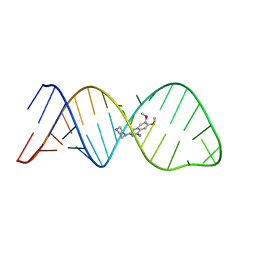 | | Solution structure of small molecule-influenza RNA complex | | Descriptor: | 6,7-dimethoxy-2-(piperazin-1-yl)quinazolin-4-amine, RNA (32-MER) | | Authors: | Lee, M.-K, Varani, G, Choi, B.-S, Pellecchia, M, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2012-08-01 | | Release date: | 2012-08-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A novel small-molecule binds to the influenza A virus RNA promoter and inhibits viral replication.
Chem.Commun.(Camb.), 50, 2014
|
|
3VRP
 
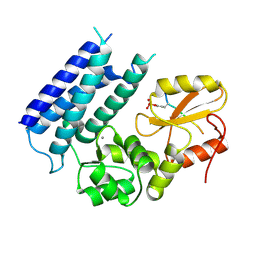 | | Crystal structure of the tyrosine kinase binding domain of Cbl-c in complex with phospho-EGFR peptide | | Descriptor: | CALCIUM ION, Epidermal growth factor receptor, Signal transduction protein CBL-C | | Authors: | Takeshita, K, Tezuka, T, Isozaki, Y, Yamashita, E, Suzuki, M, Yamanashi, Y, Yamamoto, T, Nakagawa, A. | | Deposit date: | 2012-04-13 | | Release date: | 2013-03-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural flexibility regulates phosphopeptide-binding activity of the tyrosine kinase binding domain of Cbl-c.
J.Biochem., 152, 2012
|
|
7UNL
 
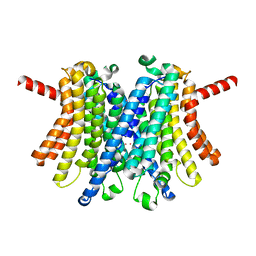 | | Human TMEM175 in an open state | | Descriptor: | Endosomal/lysosomal potassium channel TMEM175, POTASSIUM ION | | Authors: | Oh, S, Hite, R.K. | | Deposit date: | 2022-04-11 | | Release date: | 2022-06-01 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Differential ion dehydration energetics explains selectivity in the non-canonical lysosomal K + channel TMEM175.
Elife, 11, 2022
|
|
7UFM
 
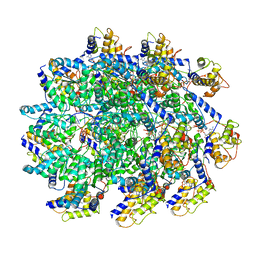 | | VchTnsC AAA+ with DNA (double heptamer) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (36-MER), VchTnsC | | Authors: | Fernandez, I.S, Sternberg, S.H. | | Deposit date: | 2022-03-22 | | Release date: | 2022-06-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Selective TnsC recruitment enhances the fidelity of RNA-guided transposition.
Nature, 609, 2022
|
|
7UFI
 
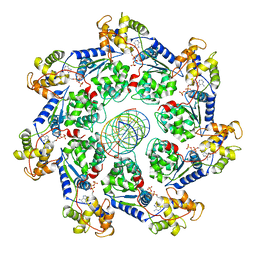 | | VchTnsC AAA+ ATPase with DNA, single heptamer | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*CP*TP*CP*CP*AP*GP*TP*AP*CP*AP*GP*CP*GP*CP*GP*GP*CP*TP*GP*AP*A)-3'), DNA (5'-D(P*TP*TP*CP*AP*GP*CP*CP*GP*CP*GP*CP*TP*GP*TP*AP*CP*TP*GP*GP*AP*G)-3'), ... | | Authors: | Fernandez, I.S, Sternberg, S.H. | | Deposit date: | 2022-03-22 | | Release date: | 2022-06-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Selective TnsC recruitment enhances the fidelity of RNA-guided transposition.
Nature, 609, 2022
|
|
7UNM
 
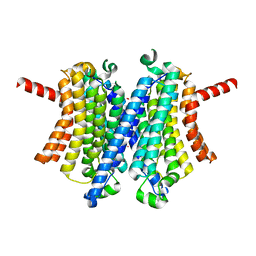 | | Human TMEM175 in an closed state | | Descriptor: | Endosomal/lysosomal potassium channel TMEM175, POTASSIUM ION | | Authors: | Oh, S, Hite, R.K. | | Deposit date: | 2022-04-11 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Differential ion dehydration energetics explains selectivity in the non-canonical lysosomal K + channel TMEM175.
Elife, 11, 2022
|
|
7F92
 
 | | Structure of connexin43/Cx43/GJA1 gap junction intercellular channel in LMNG/CHS detergents at pH ~8.0 | | Descriptor: | Gap junction alpha-1 protein, TETRADECANE | | Authors: | Lee, H.J, Cha, H.J, Jeong, H, Lee, S.N, Lee, C.W, Woo, J.S. | | Deposit date: | 2021-07-03 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM.
Nat Commun, 14, 2023
|
|
7F93
 
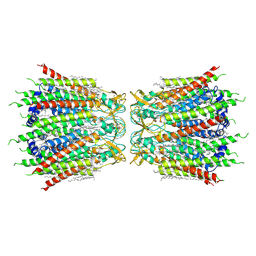 | | Structure of connexin43/Cx43/GJA1 gap junction intercellular channel in nanodiscs with soybean lipids at pH ~8.0 | | Descriptor: | Gap junction alpha-1 protein, TETRADECANE | | Authors: | Lee, H.J, Cha, H.J, Jeong, H, Lee, S.N, Lee, C.W, Woo, J.S. | | Deposit date: | 2021-07-03 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM.
Nat Commun, 14, 2023
|
|
