4J76
 
 | | Crystal Structure of a parasite tRNA synthetase, ligand-free | | Descriptor: | GLYCEROL, Tryptophanyl-tRNA synthetase | | Authors: | Koh, C.Y, Kim, J.E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2013-02-12 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal structures of Plasmodium falciparum cytosolic tryptophanyl-tRNA synthetase and its potential as a target for structure-guided drug design.
Mol.Biochem.Parasitol., 189, 2013
|
|
9KK0
 
 | | Solution structure of kappa-conotoxin RIIIJ | | Descriptor: | Kappa-conotoxin RIIIJ | | Authors: | Lushpa, V.A, Vassilevski, A.A, Bocharov, E.V, Olivera, B.M, Rivier, J.E, Raghuraman, S. | | Deposit date: | 2024-11-12 | | Release date: | 2024-12-18 | | Method: | SOLUTION NMR | | Cite: | Solution structure of kappa-conotoxin RIIIJ
To Be Published
|
|
4I2X
 
 | | Crystal structure of Signal Regulatory Protein gamma (SIRP-gamma) in complex with FabOX117 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, FabOX117 heavy chain, ... | | Authors: | Nettleship, J.E, Ren, J, Stuart, D.I, Owens, R.J, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2012-11-23 | | Release date: | 2013-12-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal structure of signal regulatory protein gamma (SIRP gamma) in complex with an antibody Fab fragment.
Bmc Struct.Biol., 13, 2013
|
|
4NFU
 
 | | Structure of the central plant immunity signaling node EDS1 in complex with its interaction partner SAG101 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, EDS1, ISOPROPYL ALCOHOL, ... | | Authors: | Wagner, S, Stuttmann, J, Rietz, S, Guerois, R, Niefind, K, Parker, J.E. | | Deposit date: | 2013-11-01 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural Basis for Signaling by Exclusive EDS1 Heteromeric Complexes with SAG101 or PAD4 in Plant Innate Immunity.
Cell Host Microbe, 14, 2013
|
|
4JCY
 
 | | Crystal structure of the Restriction-Modification Controller Protein C.Csp231I OR operator complex | | Descriptor: | Csp231I C protein, DNA (5'-D(*AP*AP*AP*CP*TP*AP*AP*GP*AP*AP*AP*AP*TP*CP*TP*TP*AP*GP*CP*AP*A)-3'), DNA (5'-D(*TP*TP*GP*CP*TP*AP*AP*GP*AP*TP*TP*TP*TP*CP*TP*TP*AP*GP*TP*TP*T)-3'), ... | | Authors: | Shevtsov, M.B, Streeter, S.D, Thresh, S.J, Mcgeehan, J.E, Kneale, G.G. | | Deposit date: | 2013-02-22 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of DNA binding by C.Csp231I, a member of a novel class of R-M controller proteins regulating gene expression.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4JGS
 
 | |
4JF2
 
 | |
4KGK
 
 | | Bacterial tRNA(HIS) Guanylyltransferase (Thg1)-Like Protein in complex with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Thg1-like uncharacterized protein | | Authors: | Hyde, S.J, Rao, B.S, Eckenroth, B.E, Jackman, J.E, Doublie, S. | | Deposit date: | 2013-04-29 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural Studies of a Bacterial tRNA(HIS) Guanylyltransferase (Thg1)-Like Protein, with Nucleotide in the Activation and Nucleotidyl Transfer Sites.
Plos One, 8, 2013
|
|
4KQO
 
 | | Crystal structure of penicillin-binding protein 3 from pseudomonas aeruginosa in complex with piperacillin | | Descriptor: | CHLORIDE ION, GLYCEROL, IMIDAZOLE, ... | | Authors: | Nettleship, J.E, Stuart, D.I, Owens, R.J, Ren, J. | | Deposit date: | 2013-05-15 | | Release date: | 2013-11-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Binding of (5S)-Penicilloic Acid to Penicillin Binding Protein 3.
Acs Chem.Biol., 8, 2013
|
|
4ITR
 
 | |
4IXQ
 
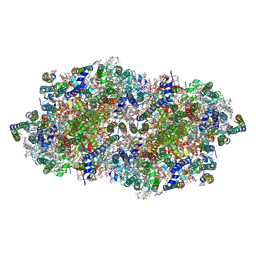 | | RT fs X-ray diffraction of Photosystem II, dark state | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Alonso-Mori, R, Tran, R, Hattne, J, Gildea, R.J, Echols, N, Gloeckner, C, Hellmich, J, Laksmono, H, Sierra, R.G, Lassalle-Kaiser, B, Koroidov, S, Lampe, A, Han, G, Gul, S, DiFiore, D, Milathianaki, D, Fry, A.R, Miahnahri, A, Schafer, D.W, Messerschmidt, M, Seibert, M.M, Koglin, J.E, Sokaras, D, Weng, T.-C, Sellberg, J, Latimer, M.J, Grosse-Kunstleve, R.W, Zwart, P.H, White, W.E, Glatzel, P, Adams, P.D, Bogan, M.J, Williams, G.J, Boutet, S, Messinger, J, Zouni, A, Sauter, N.K, Yachandra, V.K, Bergmann, U, Yano, J. | | Deposit date: | 2013-01-27 | | Release date: | 2013-02-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (5.7 Å) | | Cite: | Simultaneous femtosecond X-ray spectroscopy and diffraction of photosystem II at room temperature.
Science, 340, 2013
|
|
4J4J
 
 | |
4IXR
 
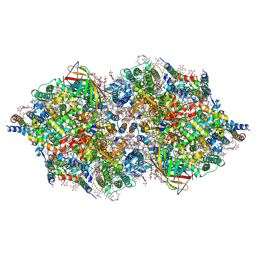 | | RT fs X-ray diffraction of Photosystem II, first illuminated state | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Alonso-Mori, R, Tran, R, Hattne, J, Gildea, R.J, Echols, N, Gloeckner, C, Hellmich, J, Laksmono, H, Sierra, R.G, Lassalle-Kaiser, B, Koroidov, S, Lampe, A, Han, G, Gul, S, DiFiore, D, Milathianaki, D, Fry, A.R, Miahnahri, A, Schafer, D.W, Messerschmidt, M, Seibert, M.M, Koglin, J.E, Sokaras, D, Weng, T.-C, Sellberg, J, Latimer, M.J, Grosse-Kunstleve, R.W, Zwart, P.H, White, W.E, Glatzel, P, Adams, P.D, Bogan, M.J, Williams, G.J, Boutet, S, Messinger, J, Zouni, A, Sauter, N.K, Yachandra, V.K, Bergmann, U, Yano, J. | | Deposit date: | 2013-01-27 | | Release date: | 2013-02-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (5.9 Å) | | Cite: | Simultaneous femtosecond X-ray spectroscopy and diffraction of photosystem II at room temperature.
Science, 340, 2013
|
|
4KQR
 
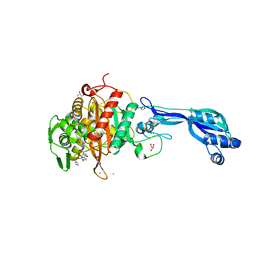 | | CRYSTAL STRUCTURE OF PENICILLIN-BINDING PROTEIN 3 FROM PSEUDOMONAS AERUGINOSA IN COMPLEX WITH (5S)-Penicilloic Acid | | Descriptor: | (2S,4S)-2-[(R)-carboxy{[(2R)-2-{[(4-ethyl-2,3-dioxopiperazin-1-yl)carbonyl]amino}-2-phenylacetyl]amino}methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nettleship, J.E, Stuart, D.I, Owens, R.J, Ren, J. | | Deposit date: | 2013-05-15 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Binding of (5S)-Penicilloic Acid to Penicillin Binding Protein 3.
Acs Chem.Biol., 8, 2013
|
|
4K8Y
 
 | | Atomic resolution crystal structures of Kallikrein-Related Peptidase 4 complexed with Sunflower Trypsin Inhibitor (SFTI-1) | | Descriptor: | Kallikrein-4, Trypsin inhibitor 1 | | Authors: | Ilyichova, O.V, Swedberg, J.E, de Veer, S.J, Sit, K.C, Harris, J.M, Buckle, A.M. | | Deposit date: | 2013-04-19 | | Release date: | 2014-04-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Direct and indirect mechanisms of KLK4 inhibition revealed by structure and dynamics
Sci Rep, 6, 2016
|
|
4KEL
 
 | | Atomic resolution crystal structure of Kallikrein-Related Peptidase 4 complexed with a modified SFTI inhibitor FCQR(N) | | Descriptor: | Kallikrein-4, Trypsin inhibitor 1 | | Authors: | Ilyichova, O.V, Swedberg, J.E, de Veer, S.J, Sit, K.C, Harris, J.M, Buckle, A.M. | | Deposit date: | 2013-04-25 | | Release date: | 2014-04-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.148 Å) | | Cite: | KLK4 Inhibition by Cyclic and Acyclic Peptides: Structural and Dynamical Insights into Standard-Mechanism Protease Inhibitors.
Biochemistry, 58, 2019
|
|
4J4Q
 
 | | Crystal structure of active conformation of GPCR opsin stabilized by octylglucoside | | Descriptor: | ACETATE ION, Guanine nucleotide-binding protein G(t) subunit alpha-1, PALMITIC ACID, ... | | Authors: | Park, J.H, Morizumi, T, Li, Y, Hong, J.E, Pai, E.F, Hofmann, K.P, Choe, H.W, Ernst, O.P. | | Deposit date: | 2013-02-07 | | Release date: | 2013-10-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Opsin, a structural model for olfactory receptors?
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
4JCX
 
 | | Crystal structure of the Restriction-Modification Controller Protein C.Csp231I OL operator complex | | Descriptor: | Csp231I C protein, DNA (5'-D(*AP*CP*AP*CP*TP*AP*AP*GP*GP*AP*AP*AP*AP*CP*TP*TP*AP*GP*TP*AP*A)-3'), DNA (5'-D(*TP*TP*AP*CP*TP*AP*AP*GP*TP*TP*TP*TP*CP*CP*TP*TP*AP*GP*TP*GP*T)-3') | | Authors: | Shevtsov, M.B, Streeter, S.D, Thresh, S.J, Mcgeehan, J.E, Kneale, G.G. | | Deposit date: | 2013-02-22 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of DNA binding by C.Csp231I, a member of a novel class of R-M controller proteins regulating gene expression.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4JQD
 
 | | Crystal structure of the Restriction-Modification Controller Protein C.Csp231I OL operator complex | | Descriptor: | Csp231I C protein, DNA (5'-D(*AP*CP*AP*CP*TP*AP*AP*GP*GP*AP*AP*AP*AP*CP*TP*TP*AP*GP*TP*AP*A)-3'), DNA (5'-D(*TP*TP*AP*CP*TP*AP*AP*GP*TP*TP*TP*TP*CP*CP*TP*TP*AP*GP*TP*GP*T)-3') | | Authors: | Shevtsov, M.B, Streeter, S.D, Thresh, S.J, McGeehan, J.E, Kneale, G.G. | | Deposit date: | 2013-03-20 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural analysis of DNA binding by C.Csp231I, a member of a novel class of R-M controller proteins regulating gene expression.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4L4R
 
 | | Structural Characterisation of the Apo-form of Human Lactate Dehydrogenase M Isozyme | | Descriptor: | L-lactate dehydrogenase A chain | | Authors: | Dempster, S, Harper, S, Moses, J.E, Dreveny, I. | | Deposit date: | 2013-06-09 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of the apo form and NADH binary complex of human lactate dehydrogenase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LI5
 
 | | EGFR-K IN COMPLEX WITH N-[3-[[5-chloro-4-(1H-indol-3-yl)pyrimidin-2-yl]amino]-4-methoxy-phenyl] Prop-2-enamide | | Descriptor: | Epidermal growth factor receptor, N-(3-{[5-chloro-4-(1H-indol-3-yl)pyrimidin-2-yl]amino}-4-methoxyphenyl)propanamide, SODIUM ION | | Authors: | Debreczeni, J.E, Seiffert, G.B, Kiefersauer, R, Augustin, M, Nagel, S, Ward, R, Anderton, M, Ashton, S, Bethel, P, Box, M, Butterworth, S, Colclough, N, Chroley, C, Chuaqui, C, Cross, D, Eberlein, C, Finlay, R, Hill, G, Grist, M, Klinowska, T, Lane, C, Martin, S, Orme, J, Smith, P, Wang, F, Waring, M. | | Deposit date: | 2013-07-02 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structure- and Reactivity-Based Development of Covalent Inhibitors of the Activating and Gatekeeper Mutant Forms of the Epidermal Growth Factor Receptor (EGFR).
J.Med.Chem., 56, 2013
|
|
4L4S
 
 | | Structural characterisation of the NADH binary complex of human lactate dehydrogenase M isozyme | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, L-lactate dehydrogenase A chain | | Authors: | Dempster, S, Harper, S, Moses, J.E, Dreveny, I. | | Deposit date: | 2013-06-09 | | Release date: | 2014-04-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural characterization of the apo form and NADH binary complex of human lactate dehydrogenase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3TIO
 
 | | Crystal structures of yrdA from Escherichia coli, a homologous protein of gamma-class carbonic anhydrase, show possible allosteric conformations | | Descriptor: | PHOSPHATE ION, Protein YrdA, ZINC ION | | Authors: | Park, H.M, Choi, J.W, Lee, J.E, Jung, C.H, Kim, B.Y, Kim, J.S. | | Deposit date: | 2011-08-21 | | Release date: | 2012-08-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structures of the gamma-class carbonic anhydrase homologue YrdA suggest a possible allosteric switch
Acta Crystallogr.,Sect.D, 68, 2012
|
|
6IAL
 
 | | Porcine E.coli heat-labile enterotoxin B-pentamer in complex with Lacto-N-neohexaose | | Descriptor: | CALCIUM ION, GLYCEROL, Heat-labile enterotoxin B chain, ... | | Authors: | Heim, J.B, Heggelund, J.E, Krengel, U. | | Deposit date: | 2018-11-27 | | Release date: | 2019-02-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Specificity ofEscherichia coliHeat-Labile Enterotoxin Investigated by Single-Site Mutagenesis and Crystallography.
Int J Mol Sci, 20, 2019
|
|
3VZL
 
 | | Crystal structure of the Bacillus circulans endo-beta-(1,4)-xylanase (BcX) N35H mutant | | Descriptor: | Endo-1,4-beta-xylanase, SULFATE ION | | Authors: | Ludwiczek, M.L, D'Angelo, I, Yalloway, G.N, Okon, M, Nielsen, J.E, Strynadka, N.C, Withers, S.G, McIntosh, L.P. | | Deposit date: | 2012-10-15 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Strategies for modulating the pH-dependent activity of a family 11 glycoside hydrolase
Biochemistry, 52, 2013
|
|
