8K4Q
 
 | |
8PHI
 
 | | Crystal structure of prefusion-stabilized RSV F Variant DS-Cav1 in complex with Lonafarnib | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 4-{2-[4-(3,10-DIBROMO-8-CHLORO-6,11-DIHYDRO-5H-BENZO[5,6]CYCLOHEPTA[1,2-B]PYRIDIN-11-YL)PIPERIDIN-1-YL]-2-OXOETHYL}PIPERIDINE-1-CARBOXAMIDE, CALCIUM ION, ... | | Authors: | Rajak, M, Krey, T. | | Deposit date: | 2023-06-19 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.289 Å) | | Cite: | Drug repurposing screen identifies lonafarnib as respiratory syncytial virus fusion protein inhibitor.
Nat Commun, 15, 2024
|
|
7BW1
 
 | | Crystal structure of Steroid 5-alpha-reductase 2 in complex with Finasteride | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-oxo-5-alpha-steroid 4-dehydrogenase 2, SULFATE ION, ... | | Authors: | Xiao, Q, Zhang, C, Wei, Z. | | Deposit date: | 2020-04-13 | | Release date: | 2020-08-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of human steroid 5 alpha-reductase 2 with anti-androgen drug finasteride.
Res Sq, 2020
|
|
5V0Y
 
 | | Solution structure of arenicin-3. | | Descriptor: | arenicin-3 | | Authors: | Edwards, I.A, Mobli, M. | | Deposit date: | 2017-02-28 | | Release date: | 2018-08-08 | | Last modified: | 2020-06-10 | | Method: | SOLUTION NMR | | Cite: | An amphipathic peptide with antibiotic activity against multidrug-resistant Gram-negative bacteria
Nat Commun, 2020
|
|
8T4Y
 
 | | Human HCN1 F186C S264C C309A bound to cAMP, reconstituted in LMNG + SPL | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 | | Authors: | Burtscher, V, Mount, J, Cowgill, J, Chang, Y, Bickel, K, Yuan, P, Chanda, B. | | Deposit date: | 2023-06-12 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural basis for hyperpolarization-dependent opening of human HCN1 channel.
Nat Commun, 15, 2024
|
|
8T4M
 
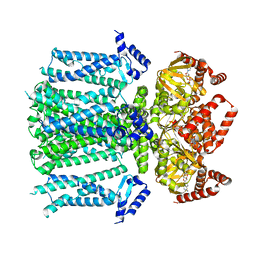 | | Closed human HCN1 F186C S264C bound to cAMP, reconstituted in LMNG + SPL | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 | | Authors: | Burtscher, V, Mount, J, Cowgill, J, Chang, Y, Bickel, K, Yuan, P, Chanda, B. | | Deposit date: | 2023-06-09 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural basis for hyperpolarization-dependent opening of human HCN1 channel.
Nat Commun, 15, 2024
|
|
8T50
 
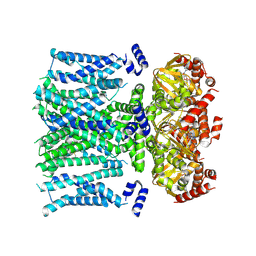 | | Open human HCN1 F186C S264C bound to cAMP, reconstituted in LMNG + SPL | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 | | Authors: | Burtscher, V, Mount, J, Cowgill, J, Chang, Y, Bickel, K, Yuan, P, Chanda, B. | | Deposit date: | 2023-06-12 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for hyperpolarization-dependent opening of human HCN1 channel.
Nat Commun, 15, 2024
|
|
7WFW
 
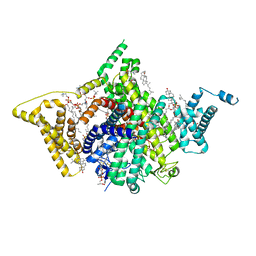 | | Apo human Nav1.8 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, N, Pan, X.J, Huang, X.S, Huang, G.X. | | Deposit date: | 2021-12-27 | | Release date: | 2022-08-03 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for high-voltage activation and subtype-specific inhibition of human Na v 1.8.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WFR
 
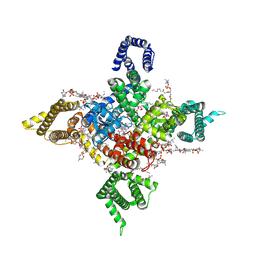 | | Human Nav1.8 with A-803467, class III | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, N, Pan, X.J, Huang, X.S, Huang, G.X. | | Deposit date: | 2021-12-27 | | Release date: | 2022-08-03 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for high-voltage activation and subtype-specific inhibition of human Na v 1.8.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WEL
 
 | | Human Nav1.8 with A-803467, class II | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, N, Pan, X.J, Huang, X.S, Huang, G.X. | | Deposit date: | 2021-12-23 | | Release date: | 2022-08-03 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for high-voltage activation and subtype-specific inhibition of human Na v 1.8.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WE4
 
 | | Human Nav1.8 with A-803467, class I | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, N, Pan, X.J, Huang, X.S, Huang, G.X. | | Deposit date: | 2021-12-22 | | Release date: | 2022-08-03 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for high-voltage activation and subtype-specific inhibition of human Na v 1.8.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8U08
 
 | |
8U03
 
 | |
8V2E
 
 | |
8TZN
 
 | |
8TZW
 
 | |
6LKC
 
 | | Crystal structure of PfaD from Shewanella piezotolerans in complex with FMN | | Descriptor: | CALCIUM ION, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Zhang, M.L, Li, Q, Meng, S.S, Guo, L.J, He, L, Huang, J.Z, Li, L, Zhang, H.D. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural Insights into the Trans -Acting Enoyl Reductase in the Biosynthesis of Long-Chain Polyunsaturated Fatty Acids in Shewanella piezotolerans .
J.Agric.Food Chem., 69, 2021
|
|
4WZF
 
 | | Crystal structural basis for Rv0315, an immunostimulatory antigen and pseudo beta-1, 3-glucanase of Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, 1,3-beta-glucanase, CALCIUM ION | | Authors: | Dong, W.Y, Fu, Z.F, Peng, G.Q. | | Deposit date: | 2014-11-19 | | Release date: | 2015-12-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Crystal structural basis for Rv0315, an immunostimulatory antigen and inactive beta-1,3-glucanase of Mycobacterium tuberculosis.
Sci Rep, 5, 2015
|
|
5F8U
 
 | |
8I5B
 
 | | Structure of human Nav1.7 in complex with bupivacaine | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, Q.R, Yan, N. | | Deposit date: | 2023-01-24 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
8I5X
 
 | | Structure of human Nav1.7 in complex with Vinpocetine | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Wu, Q.R, Yan, N. | | Deposit date: | 2023-01-26 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
8I5Y
 
 | | Structure of human Nav1.7 in complex with vixotrigine | | Descriptor: | (2S,3R,4E)-2-(acetylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Wu, Q.R, Yan, N. | | Deposit date: | 2023-01-26 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
8I5G
 
 | | Structure of human Nav1.7 in complex with PF-05089771 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Wu, Q.R, Yan, N. | | Deposit date: | 2023-01-25 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
8J4F
 
 | | Structure of human Nav1.7 in complex with Hardwickii acid | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, (4~{a}~{R},5~{S},6~{R},8~{a}~{R})-5-[2-(furan-3-yl)ethyl]-5,6,8~{a}-trimethyl-3,4,4~{a},6,7,8-hexahydronaphthalene-1-carboxylic acid, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Wu, Q.R, Yan, N. | | Deposit date: | 2023-04-19 | | Release date: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
8YLB
 
 | | Cocrystal structures of agonists compound 1 with HsClpP | | Descriptor: | 5-[(2-methylphenyl)methyl]-11-(phenylmethyl)-2,5,7,11-tetrazatricyclo[7.4.0.0^{2,6}]trideca-1(9),6-dien-8-one, ATP-dependent Clp protease proteolytic subunit, mitochondrial | | Authors: | Zhao, N, Zhu, Y, Bao, R. | | Deposit date: | 2024-03-06 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Rational Design of a Novel Class of Human ClpP Agonists through a Ring-Opening Strategy with Enhanced Antileukemia Activity.
J.Med.Chem., 67, 2024
|
|
