2LCB
 
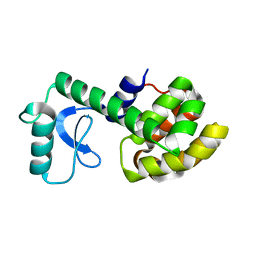 | | Solution Structure of a Minor and Transiently Formed State of a T4 Lysozyme Mutant | | Descriptor: | Lysozyme | | Authors: | Bouvignies, G, Vallurupalli, P, Hansen, D, Correia, B, Lange, O, Bah, A, Vernon, R.M, Dahlquist, F.W, Baker, D, Kay, L.E. | | Deposit date: | 2011-04-26 | | Release date: | 2011-08-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a minor and transiently formed state of a T4 lysozyme mutant.
Nature, 477, 2011
|
|
5A0E
 
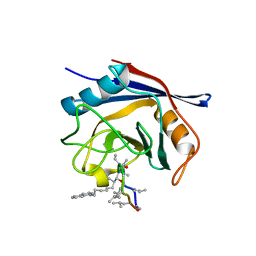 | | Crystal structure of cyclophilin D in complex with CsA analogue, JW47. | | Descriptor: | JW47, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE F, MITOCHONDRIAL | | Authors: | Warne, J, Pryce, G, Hill, J, Shi, X, Lenneras, F, Puentes, F, Kip, M, Hilditch, L, Walker, P, Simone, M, Chan, A.W.E, Towers, G, Coker, A.R, Duchen, M, Szabadkai, G, Baker, D, Selwood, D.L. | | Deposit date: | 2015-04-19 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Selective Inhibition of the Mitochondrial Permeability Transition Pore Protects Against Neuro-Degeneration in Experimental Multiple Sclerosis.
J.Biol.Chem., 291, 2016
|
|
2LPZ
 
 | | Atomic model of the Type-III Secretion System Needle | | Descriptor: | Protein prgI | | Authors: | Loquet, A, Sgourakis, N.G, Gupta, R, Giller, K, Riedel, D, Goosmann, C, Griesinger, C, Kolbe, M.G, Baker, D, Becker, S, Lange, A. | | Deposit date: | 2012-02-21 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic model of the type III secretion system needle.
Nature, 486, 2012
|
|
7M0Q
 
 | |
7M5T
 
 | |
7MWQ
 
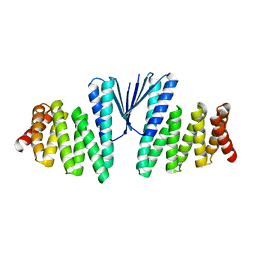 | | Structure of De Novo designed beta sheet heterodimer LHD29A53/B53 | | Descriptor: | LHD29A53, LHD29B53 | | Authors: | Bera, A.K, Sahtoe, D.D, Kang, A, Praetorius, F, Baker, D. | | Deposit date: | 2021-05-17 | | Release date: | 2022-01-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Reconfigurable asymmetric protein assemblies through implicit negative design.
Science, 375, 2022
|
|
7MWR
 
 | | Structure of De Novo designed beta sheet heterodimer LHD101A53/B4 | | Descriptor: | LHD101A54, LHD101B4, MALONATE ION | | Authors: | Bera, A.K, Sahtoe, D.D, Kang, A, Praetorius, F, Baker, D. | | Deposit date: | 2021-05-17 | | Release date: | 2022-01-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reconfigurable asymmetric protein assemblies through implicit negative design.
Science, 375, 2022
|
|
2LC9
 
 | | Solution Structure of a Minor and Transiently Formed State of a T4 Lysozyme Mutant | | Descriptor: | Lysozyme | | Authors: | Bouvignies, G, Vallurupalli, P, Hansen, D, Correia, B, Lange, O, Bah, A, Vernon, R.M, Dahlquist, F.W, Baker, D, Kay, L.E. | | Deposit date: | 2011-04-26 | | Release date: | 2011-08-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a minor and transiently formed state of a T4 lysozyme mutant.
Nature, 477, 2011
|
|
4JBC
 
 | | Crystal Structure of the computationally designed serine hydrolase 3mmj_2, Northeast Structural Genomics Consortium (NESG) Target OR318 | | Descriptor: | PHOSPHATE ION, designed serine hydrolase 3mmj_2 | | Authors: | Kuzin, A, Lew, S, Rajagopalan, S, Seetharaman, J, Maglaqui, M, Xiao, R, Lee, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Baker, D, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-02-19 | | Release date: | 2013-03-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Crystal Structure of the computationally designed serine hydrolase 3mmj_2, Northeast Structural Genomics Consortium (NESG) Target OR318
To be Published
|
|
4K0C
 
 | | Crystal Structure of the computationally designed serine hydrolase. Northeast Structural Genomics Consortium (NESG) Target OR317 | | Descriptor: | designed serine hydrolase | | Authors: | Kuzin, A, Lew, S, Rajagopalan, S, Seetharaman, J, Maglaqui, M, Xiao, R, Lee, D, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-04-03 | | Release date: | 2013-04-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Northeast Structural Genomics Consortium Target OR317
To be Published
|
|
3Q2D
 
 | | Optimization of the in silico designed Kemp eliminase KE70 by computational design and directed evolution | | Descriptor: | 5-nitro-1H-benzotriazole, Deoxyribose phosphate aldolase | | Authors: | Khersonsky, O, Rothlisberge, D, Wollacott, A.M, Murphy, P, Dym, O, Albeck, S, Kiss, G, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2010-12-20 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Optimization of the in-silico-designed kemp eliminase KE70 by computational design and directed evolution
J.Mol.Biol., 407, 2011
|
|
4JXI
 
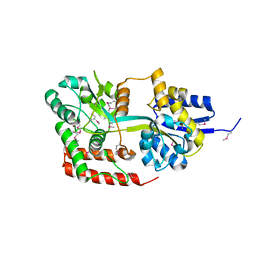 | | Directed evolution and rational design of a de novo designed esterase toward improved catalysis. Northeast Structural Genomics Consortium (NESG) Target OR184 | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, BROMIDE ION, GLYCEROL, ... | | Authors: | Kuzin, A, Smith, M.D, Richter, F, Lew, S, Seetharaman, R, Bryan, C, Lech, Z, Kiss, G, Moretti, R, Maglaqui, M, Xiao, R, Kohan, E, Smith, M, Everett, J.K, Nguyen, R, Pande, V, Hilvert, D, Kornhaber, G, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-03-28 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Directed evolution and rational design of a de novo designed esterase toward improved catalysis. Northeast Structural Genomics Consortium (NESG) Target OR184
To be Published
|
|
4KYB
 
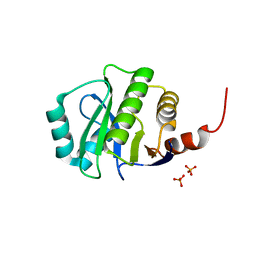 | | Crystal Structure of de novo designed serine hydrolase OSH55.14_E3, Northeast Structural Genomics Consortium Target OR342 | | Descriptor: | Designed Protein OR342, PHOSPHATE ION | | Authors: | Kuzin, A, Lew, S, Rajagopalan, S, Seetharaman, J, Mao, L, Xiao, R, Lee, D, Raja, S, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-05-28 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.909 Å) | | Cite: | Northeast Structural Genomics Consortium Target OR342
To be Published
|
|
4JGK
 
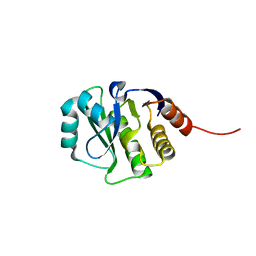 | | Crystal Structure of the evolved variant of the computationally designed serine hydrolase, Northeast Structural Genomics Consortium (NESG) Target OR275 | | Descriptor: | evolved variant of a designed serine hydrolase | | Authors: | Kuzin, A, Lew, S, Rajagopalan, S, Seetharaman, J, Mao, L, Xiao, R, Lee, D, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-03-01 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.883 Å) | | Cite: | Crystal Structure of the evolved variant of the computationally designed serine hydrolase, Northeast Structural Genomics Consortium (NESG) Target OR275
To be Published
|
|
2A3J
 
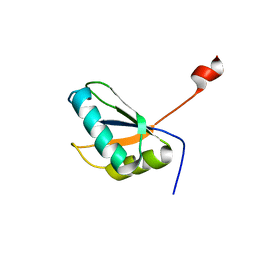 | | Structure of URNdesign, a complete computational redesign of human U1A protein | | Descriptor: | U1 small nuclear ribonucleoprotein A | | Authors: | Varani, G, Dobson, N, Dantas, G, Baker, D. | | Deposit date: | 2005-06-24 | | Release date: | 2006-06-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Structural Validation of the Computational Redesign of Human U1A Protein
Structure, 14, 2006
|
|
4J4Z
 
 | | Crystal structure of the improved variant of the evolved serine hydrolase, OSH55.4_H1.2, bond with sulfate ion in the active site, Northeast Structural Genomics Consortium (NESG) Target OR301 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, Designed serine hydrolase variant OSH55.4_H1.2, ... | | Authors: | Kuzin, A.P, Lew, S, Rajagopalan, S, Maglaqui, M, Xiao, R, Lee, D, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-02-07 | | Release date: | 2013-03-06 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystal structure of the improved variant of the evolved serine hydrolase, OSH55.4_H1.2, bond with sulfate ion in the active site, Northeast Structural Genomics Consortium (NESG) Target OR301
To be Published
|
|
2L69
 
 | | Solution NMR Structure of de novo designed protein, P-loop NTPase fold, Northeast Structural Genomics Consortium Target OR28 | | Descriptor: | Rossmann 2x3 fold protein | | Authors: | Liu, G, Koga, N, Koga, R, Xiao, R, Mao, A, Mao, B, Patel, D, Ciccosanti, C, Hamilton, K, Acton, T.B, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-11-17 | | Release date: | 2011-01-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of de novo designed rossmann 2x3 fold protein, Northeast Structural Genomics Consortium Target OR28
To be Published
|
|
3TC6
 
 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR63. | | Descriptor: | DI(HYDROXYETHYL)ETHER, Indole-3-glycerol phosphate synthase, PHOSPHATE ION | | Authors: | Vorobiev, S, Su, M, Bjelic, S, Kipnis, Y, Wang, L, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-08-08 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Exploration of alternate catalytic mechanisms and optimization strategies for retroaldolase design.
J.Mol.Biol., 426, 2014
|
|
2ERH
 
 | | Crystal Structure of the E7_G/Im7_G complex; a designed interface between the colicin E7 DNAse and the Im7 immunity protein | | Descriptor: | Colicin E7, Colicin E7 immunity protein | | Authors: | Joachimiak, L.A, Kortemme, T, Stoddard, B.L, Baker, D. | | Deposit date: | 2005-10-24 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computational Design of a New Hydrogen Bond Network and at Least a 300-fold Specificity Switch at a Protein-Protein Interface.
J.Mol.Biol., 361, 2006
|
|
3TC7
 
 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR62. | | Descriptor: | ACETIC ACID, Indole-3-glycerol phosphate synthase, PHOSPHATE ION | | Authors: | Vorobiev, S, Su, M, Bjelic, S, Kipnis, Y, Wang, L, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-08-08 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Exploration of alternate catalytic mechanisms and optimization strategies for retroaldolase design.
J.Mol.Biol., 426, 2014
|
|
2MRA
 
 | | Solution NMR Structure of De novo designed protein, Northeast Structural Genomics Consortium (NESG) Target OR459 | | Descriptor: | De novo designed protein OR459 | | Authors: | Pulavarti, S.V.S.R.K, Kipnis, Y, Sukumaran, D, Maglaqui, M, Janjua, H, Mao, L, Xiao, R, Kornhaber, G, Baker, D, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-07-02 | | Release date: | 2014-09-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of De novo designed protein, Northeast Structural Genomics Consortium (NESG) Target OR459
To be Published
|
|
5TX8
 
 | |
5TPJ
 
 | | Crystal structure of a de novo designed protein with curved beta-sheet | | Descriptor: | denovo NTF2 | | Authors: | Basanta, B, Oberdorfer, G, Marcos, E, Chidyausiku, T.M, Sankaran, B, Baker, D. | | Deposit date: | 2016-10-20 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
4OYD
 
 | | Crystal structure of a computationally designed inhibitor of an Epstein-Barr viral Bcl-2 protein | | Descriptor: | 1,2-ETHANEDIOL, Apoptosis regulator BHRF1, Computationally designed Inhibitor | | Authors: | Shen, B, Procko, E, Baker, D, Stoddard, B. | | Deposit date: | 2014-02-11 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A computationally designed inhibitor of an epstein-barr viral bcl-2 protein induces apoptosis in infected cells.
Cell, 157, 2014
|
|
5U35
 
 | | Crystal structure of a de novo designed protein with curved beta-sheet | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-METHOXYETHANOL, CHLORIDE ION, ... | | Authors: | Oberdorfer, G, Marcos, E, Basanta, B, Chidyausiku, T.M, Sankaran, B, Zwart, P.H, Baker, D. | | Deposit date: | 2016-12-01 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
