6S7U
 
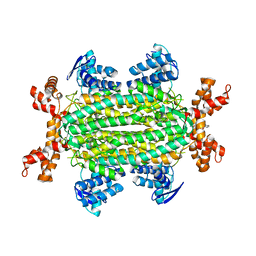 | | Fumarate hydratase of Mycobacterium tuberculosis in complex with formate and allosteric modulator N-(5-(Azepan-1-ylsulfonyl)-2-methoxyphenyl)-2-(1H-indol-3-yl)acetamide | | Descriptor: | FORMIC ACID, Fumarate hydratase class II, MAGNESIUM ION, ... | | Authors: | Whitehouse, A.J, Libardo, M.D, Kasbekar, M, Brear, P, Fischer, G, Thomas, C.J, Barry, C.E, Boshoff, H.I, Coyne, A.G, Abell, C. | | Deposit date: | 2019-07-06 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Targeting of Fumarate Hydratase fromMycobacterium tuberculosisUsing Allosteric Inhibitors with a Dimeric-Binding Mode.
J.Med.Chem., 62, 2019
|
|
8C8S
 
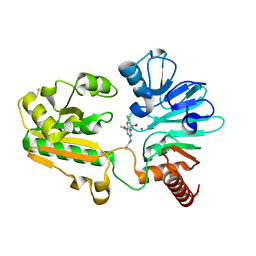 | | Crystal structure of human DNA cross-link repair 1A in complex with hydroxamic acid inhibitor (compound 21). | | Descriptor: | (2~{R})-3-[6-chloranyl-2-(prop-2-enylamino)quinazolin-4-yl]-2-methyl-~{N}-oxidanyl-propanamide, DNA cross-link repair 1A protein, ZINC ION | | Authors: | Yosaatmadja, Y, Newman, J.A, Baddock, H.T, Bielinski, M, von Delft, F, Bountra, C, McHugh, P.J, Schofield, C.J, Gileadi, O. | | Deposit date: | 2023-01-20 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human DNA cross-link repair 1A in complex with hydroxamic acid inhibitor (compound 21).
To Be Published
|
|
6S87
 
 | |
6RFP
 
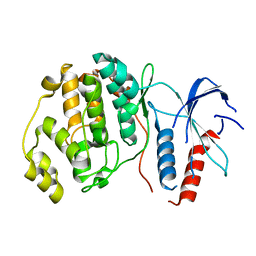 | | ERK2 MAP kinase with mutations at Helix-G | | Descriptor: | Mitogen-activated protein kinase 1 | | Authors: | Livnah, O, Eitan-Wexler, M. | | Deposit date: | 2019-04-16 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The bacterial metalloprotease NleD selectively cleaves mitogen-activated protein kinases that have high flexibility in their activation loop.
J.Biol.Chem., 295, 2020
|
|
6RGE
 
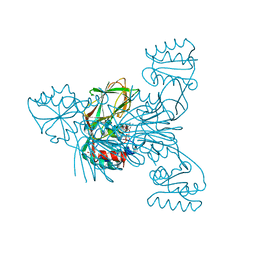 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with an inhibitor | | Descriptor: | CITRIC ACID, NAD kinase 1, [(2~{R},3~{R},4~{R},5~{R})-2-[8-[3-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy]prop-1-ynyl]-6-azanyl-purin-9-yl]-5-(hydroxymethyl)-4-oxidanyl-oxolan-3-yl] dihydrogen phosphate | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2019-04-16 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with an inhibitor
To Be Published
|
|
6S6D
 
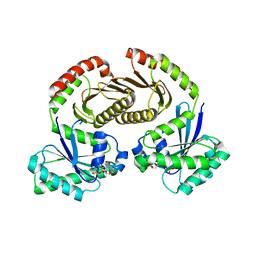 | | Crystal structure of RagA-Q66L-GTP/RagC-S75N-GDP GTPase heterodimer complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Anandapadamanaban, M, Masson, G.R, Perisic, O, Kaufman, J, Williams, R.L. | | Deposit date: | 2019-07-02 | | Release date: | 2019-10-16 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Architecture of human Rag GTPase heterodimers and their complex with mTORC1.
Science, 366, 2019
|
|
6RLR
 
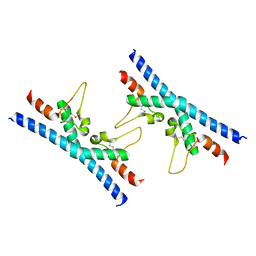 | | Crystal structure of CD9 large extracellular loop | | Descriptor: | CD9 antigen | | Authors: | Neviani, V, Kroon-Batenburg, L, Lutz, M, Pearce, N.M, Pos, W, Schotte, R, Spits, H, Gros, P. | | Deposit date: | 2019-05-02 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Implications for tetraspanin-enriched microdomain assembly based on structures of CD9 with EWI-F.
Life Sci Alliance, 3, 2020
|
|
6S6U
 
 | |
6RGD
 
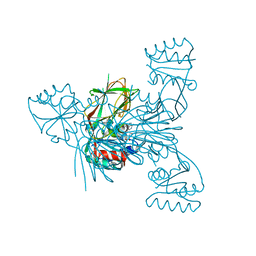 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complexe with an inhibitor | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[[4-[6-azanyl-9-[(2~{R},3~{R},4~{S},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]purin-8-yl]but-3-ynylamino]methyl]oxolane-3,4-diol, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2019-04-16 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | From Substrate to Fragments to Inhibitor ActiveIn VivoagainstStaphylococcus aureus.
Acs Infect Dis., 6, 2020
|
|
6RKG
 
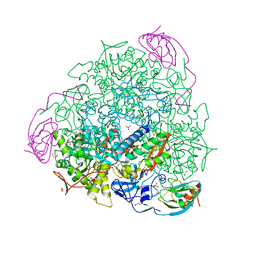 | | 1.32 A RESOLUTION OF SPOROSARCINA PASTEURII UREASE INHIBITED IN THE PRESENCE OF NBPTO AT pH 7.5 | | Descriptor: | 1,2-ETHANEDIOL, DIAMIDOPHOSPHATE, NICKEL (II) ION, ... | | Authors: | Mazzei, L, Cianci, M, Benini, S, Ciurli, S. | | Deposit date: | 2019-04-30 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | The Impact of pH on Catalytically Critical Protein Conformational Changes: The Case of the Urease, a Nickel Enzyme.
Chemistry, 25, 2019
|
|
6RLX
 
 | |
6RQ4
 
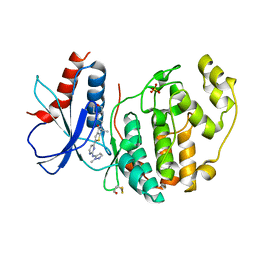 | | Inhibitor of ERK2 | | Descriptor: | 6,6-dimethyl-2-[2-[(2-methylpyrazol-3-yl)amino]pyrimidin-4-yl]-5-(2-morpholin-4-ylethyl)thieno[2,3-c]pyrrol-4-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | O'Reilly, M. | | Deposit date: | 2019-05-15 | | Release date: | 2019-12-04 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Dual-Mechanism ERK1/2 Inhibitors Exploit a Distinct Binding Mode to Block Phosphorylation and Nuclear Accumulation of ERK1/2.
Mol.Cancer Ther., 19, 2020
|
|
6RRE
 
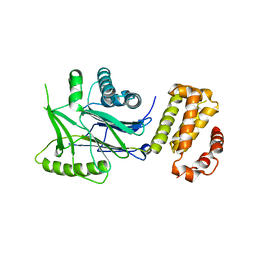 | | SidD, deAMPylase from Legionella pneumophila | | Descriptor: | Adenosine monophosphate-protein hydrolase SidD, MAGNESIUM ION | | Authors: | Tascon, I, Lucas, M, Rojas, A.L, Hierro, A. | | Deposit date: | 2019-05-17 | | Release date: | 2020-10-07 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.586 Å) | | Cite: | Structural insight into the membrane targeting domain of the Legionella deAMPylase SidD.
Plos Pathog., 16, 2020
|
|
6RX9
 
 | |
6RZE
 
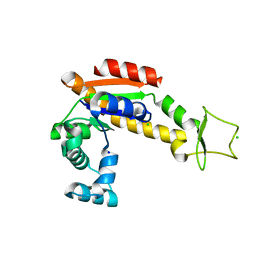 | | Crystal structure of E. coli Adenylate kinase R119A mutant | | Descriptor: | Adenylate kinase, CHLORIDE ION, SODIUM ION | | Authors: | Grundstrom, C, Rogne, P, Wolf-Watz, M, Sauer-Eriksson, A.E. | | Deposit date: | 2019-06-13 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Nucleation of an Activating Conformational Change by a Cation-pi Interaction.
Biochemistry, 58, 2019
|
|
6RYR
 
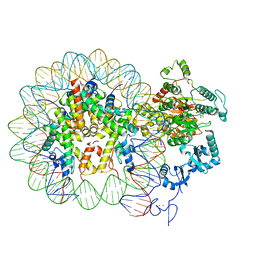 | | Nucleosome-CHD4 complex structure (single CHD4 copy) | | Descriptor: | Chromodomain-helicase-DNA-binding protein 4,Chromodomain-helicase-DNA-binding protein 4,Chromodomain-helicase-DNA-binding protein 4, DNA (149-MER), Histone H2A type 1, ... | | Authors: | Farnung, L, Ochmann, M, Cramer, P. | | Deposit date: | 2019-06-11 | | Release date: | 2020-07-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Nucleosome-CHD4 chromatin remodeller structure maps human disease mutations.
Elife, 9, 2020
|
|
6S15
 
 | | Pyridine derivative of the natural alkaloid Berberine as Human Telomeric G-quadruplex Binder | | Descriptor: | Berberine, DNA TAGGGTTAGGGT, POTASSIUM ION | | Authors: | Ferraroni, M, Bazzicalupi, C, Gratteri, P, Papi, F. | | Deposit date: | 2019-06-18 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Pyridine Derivative of the Natural Alkaloid Berberine as Human Telomeric G4-DNA Binder: A Solution and Solid-State Study.
Acs Med.Chem.Lett., 11, 2020
|
|
7UFV
 
 | | Crystal structure of the WDR domain of human DCAF1 in complex with OICR-6766 | | Descriptor: | (3P)-N-[(1S)-3-amino-1-(3-chlorophenyl)-3-oxopropyl]-3-(2-fluorophenyl)-1H-pyrazole-4-carboxamide, DDB1- and CUL4-associated factor 1, UNKNOWN ATOM OR ION | | Authors: | Kimani, S, Li, A, Li, Y, Dong, A, Hutchinson, A, Seitova, A, Wilson, B, Al-Awar, R, Vedadi, M, Brown, P, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-03-23 | | Release date: | 2022-05-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Nanomolar DCAF1 Small Molecule Ligands.
J.Med.Chem., 66, 2023
|
|
9EUU
 
 | | Structure of recombinant alpha-synuclein fibrils 1B capable of seeding GCIs in vivo | | Descriptor: | Alpha-synuclein | | Authors: | Burger, D, Kashyrina, M, Lewis, A, De Nuccio, F, Mohammed, I, de La Seigliere, H, van den Heuvel, L, Feuillie, C, Verchere, J, Berbon, M, Arotcarena, M, Retailleau, A, Bezard, E, Laferriere, F, Loquet, A, Bousset, L, Baron, T, Lofrumento, D.D, De Giorgi, F, Stahlberg, H, Ichas, F. | | Deposit date: | 2024-03-28 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (1.93 Å) | | Cite: | Multiple System Atrophy: Insights from aSyn Fibril Structure
To Be Published
|
|
7O7D
 
 | | Crystal structure of rsEGFP2 mutant V151A in the fluorescent on-state determined by synchrotron radiation at 100K | | Descriptor: | Green fluorescent protein | | Authors: | Woodhouse, J, Adam, V, Hadjidemetriou, K, Colletier, J.-P, Weik, M. | | Deposit date: | 2021-04-13 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Rational Control of Off-State Heterogeneity in a Photoswitchable Fluorescent Protein Provides Switching Contrast Enhancement.
Chemphyschem, 23, 2022
|
|
7O7U
 
 | | Crystal structure of rsEGFP2 in the non-fluorescent off-state determined by serial femtosecond crystallography at room temperature | | Descriptor: | Green fluorescent protein | | Authors: | Hadjidemetriou, K, Woodhouse, J, Coquelle, N, Barends, T.R.M, Schlichting, I, Weik, M, Colletier, J.-P. | | Deposit date: | 2021-04-13 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational Control of Off-State Heterogeneity in a Photoswitchable Fluorescent Protein Provides Switching Contrast Enhancement.
Chemphyschem, 23, 2022
|
|
7O7E
 
 | | Crystal structure of rsEGFP2 mutant V151L in the fluorescent on-state determined by synchrotron radiation at 100K | | Descriptor: | Green fluorescent protein | | Authors: | Woodhouse, J, Adam, V, Hadjidemetriou, K, Colletier, J.-P, Weik, M. | | Deposit date: | 2021-04-13 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rational Control of Off-State Heterogeneity in a Photoswitchable Fluorescent Protein Provides Switching Contrast Enhancement.
Chemphyschem, 23, 2022
|
|
7O7W
 
 | | Crystal structure of rsEGFP2 mutant V151L in the non-fluorescent off-state the determined by serial femtosecond crystallography at room temperature | | Descriptor: | Green fluorescent protein | | Authors: | Hadjidemetriou, K, Coquelle, N, Barends, T.R.M, Schlichting, I, Colletier, J.-P, Weik, M. | | Deposit date: | 2021-04-13 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Rational Control of Off-State Heterogeneity in a Photoswitchable Fluorescent Protein Provides Switching Contrast Enhancement.
Chemphyschem, 23, 2022
|
|
7O7V
 
 | | Crystal structure of rsEGFP2 mutant V151A in the fluorescent on-state determined by serial femtosecond crystallography at room temperature | | Descriptor: | Green fluorescent protein | | Authors: | Hadjidemetriou, K, Coquelle, N, Barends, T.R.M, Schlichting, I, Colletier, J.-P, Weik, M. | | Deposit date: | 2021-04-13 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rational Control of Off-State Heterogeneity in a Photoswitchable Fluorescent Protein Provides Switching Contrast Enhancement.
Chemphyschem, 23, 2022
|
|
7O7H
 
 | | Crystal structure of rsEGFP2 mutant V151L in the non-fluorescent off-state determined by synchrotron radiation at 100K | | Descriptor: | Green fluorescent protein | | Authors: | Woodhouse, J, Adam, V, Hadjidemetriou, K, Colletier, J.P, Weik, M. | | Deposit date: | 2021-04-13 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational Control of Off-State Heterogeneity in a Photoswitchable Fluorescent Protein Provides Switching Contrast Enhancement.
Chemphyschem, 23, 2022
|
|
