5ZTH
 
 | | Crystal structure of spRlmCD with U1939loop RNA at 3.24 angstrom | | Descriptor: | RNA (5'-R(P*AP*AP*AP*(MUM)P*UP*CP*CP*U)-3'), S-ADENOSYL-L-HOMOCYSTEINE, Uncharacterized RNA methyltransferase SP_1029 | | Authors: | Jiang, Y.Y, Yu, H.L. | | Deposit date: | 2018-05-03 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Unveiling the structural features that determine the dual methyltransferase activities of Streptococcus pneumoniae RlmCD
PLoS Pathog., 14, 2018
|
|
8TLR
 
 | | Crystal Structure of human HRAS G12C covalently bound to AMG 510 | | Descriptor: | AMG 510 (bound form), GTPase HRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Morstein, J, Guiley, K.Z, Shokat, K.M. | | Deposit date: | 2023-07-27 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.70003951 Å) | | Cite: | Targeting Ras-, Rho-, and Rab-family GTPases via a conserved cryptic pocket.
Cell, 2024
|
|
8UB6
 
 | |
7EAK
 
 | | Echovirus3 full particle in complex with 5G3 fab | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Feng, R. | | Deposit date: | 2021-03-07 | | Release date: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for neutralization of an anicteric hepatitis associated echovirus by a potent neutralizing antibody.
Cell Discov, 7, 2021
|
|
7EAJ
 
 | | Echovirus3 empty expanded particle in complex with 5G3 fab | | Descriptor: | VP0, VP1, VP3, ... | | Authors: | Feng, R. | | Deposit date: | 2021-03-07 | | Release date: | 2021-07-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for neutralization of an anicteric hepatitis associated echovirus by a potent neutralizing antibody.
Cell Discov, 7, 2021
|
|
7EAH
 
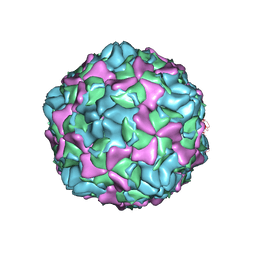 | | Echovirus3 empty expanded particle | | Descriptor: | Capsid protein VP0, Capsid protein VP1, Capsid protein VP3 | | Authors: | Feng, R. | | Deposit date: | 2021-03-07 | | Release date: | 2021-07-14 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for neutralization of an anicteric hepatitis associated echovirus by a potent neutralizing antibody.
Cell Discov, 7, 2021
|
|
7EAI
 
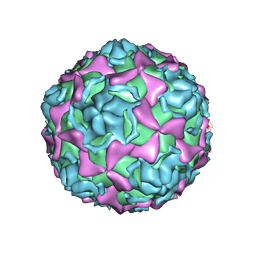 | | Echovirus3 empty compacted particle | | Descriptor: | Capsid protein VP0, Capsid protein VP1, Capsid protein VP3, ... | | Authors: | Feng, R. | | Deposit date: | 2021-03-07 | | Release date: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for neutralization of an anicteric hepatitis associated echovirus by a potent neutralizing antibody.
Cell Discov, 7, 2021
|
|
8WGH
 
 | | Cryo-EM structure of the red-shifted Fittonia albivenis PSI-LHCI | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Huang, G.Q, Li, X.X, Sui, S.F, Qin, X.C. | | Deposit date: | 2023-09-21 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structure of the red-shifted Fittonia albivenis photosystem I.
Nat Commun, 15, 2024
|
|
7CRH
 
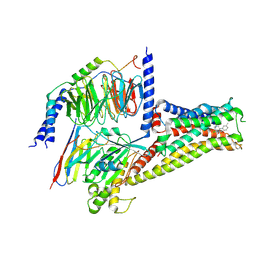 | | Cryo-EM structure of SKF83959 bound dopamine receptor DRD1-Gs signaling complex | | Descriptor: | (1S)-6-chloranyl-3-methyl-1-(3-methylphenyl)-1,2,4,5-tetrahydro-3-benzazepine-7,8-diol, D(1A) dopamine receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Yan, W, Shao, Z.H. | | Deposit date: | 2020-08-13 | | Release date: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Ligand recognition and allosteric regulation of DRD1-Gs signaling complexes.
Cell, 184, 2021
|
|
7CKY
 
 | | Cryo-EM structure of PW0464 bound dopamine receptor DRD1-Gs signaling complex | | Descriptor: | 6-[4-[3-[bis(fluoranyl)methoxy]pyridin-2-yl]oxy-2-methyl-phenyl]-1,5-dimethyl-pyrimidine-2,4-dione, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Yan, W, Shao, Z. | | Deposit date: | 2020-07-20 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Ligand recognition and allosteric regulation of DRD1-Gs signaling complexes.
Cell, 184, 2021
|
|
7CKW
 
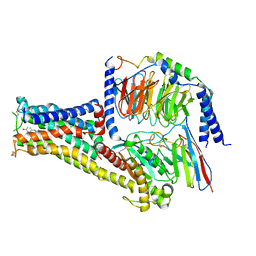 | | Cryo-EM structure of Fenoldopam bound dopamine receptor DRD1-Gs signaling complex | | Descriptor: | (1R)-6-chloranyl-1-(4-hydroxyphenyl)-2,3,4,5-tetrahydro-1H-3-benzazepine-7,8-diol, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Yan, W, Shao, W. | | Deposit date: | 2020-07-20 | | Release date: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Ligand recognition and allosteric regulation of DRD1-Gs signaling complexes.
Cell, 184, 2021
|
|
7CKX
 
 | | Cryo-EM structure of A77636 bound dopamine receptor DRD1-Gs signaling complex | | Descriptor: | (1R,3S)-3-(1-adamantyl)-1-(aminomethyl)-3,4-dihydro-1H-isochromene-5,6-diol, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Yan, W, Shao, Z. | | Deposit date: | 2020-07-20 | | Release date: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Ligand recognition and allosteric regulation of DRD1-Gs signaling complexes.
Cell, 184, 2021
|
|
7CKZ
 
 | | Cryo-EM structure of Dopamine and LY3154207 bound dopamine receptor DRD1-Gs signaling complex | | Descriptor: | 2-[2,6-bis(chloranyl)phenyl]-1-[(1S,3R)-3-(hydroxymethyl)-1-methyl-5-(3-methyl-3-oxidanyl-butyl)-3,4-dihydro-1H-isoquinolin-2-yl]ethanone, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Yan, W, Shao, Z. | | Deposit date: | 2020-07-20 | | Release date: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ligand recognition and allosteric regulation of DRD1-Gs signaling complexes.
Cell, 184, 2021
|
|
7XB2
 
 | |
7WL3
 
 | | CVB5 expended empty particle | | Descriptor: | Capsid protein, Genome polyprotein | | Authors: | Yang, P, Wang, K. | | Deposit date: | 2022-01-12 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Atomic Structures of Coxsackievirus B5 Provide Key Information on Viral Evolution and Survival.
J.Virol., 96, 2022
|
|
2JOO
 
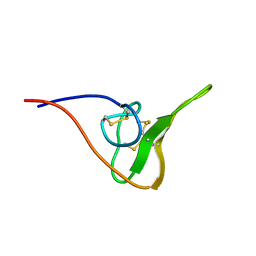 | | The NMR Solution Structure of Recombinant RGD-hirudin | | Descriptor: | Hirudin variant-1 | | Authors: | Song, X, Mo, W, Liu, X, Yan, X, Song, H, Dai, L. | | Deposit date: | 2007-03-14 | | Release date: | 2008-03-18 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of recombinant RGD-hirudin
Biochem.Biophys.Res.Commun., 360, 2007
|
|
7C34
 
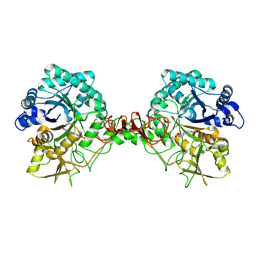 | |
8IJA
 
 | | Cryo-EM structure of human HCAR2-Gi complex with niacin | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Pan, X, Fang, Y. | | Deposit date: | 2023-02-26 | | Release date: | 2024-01-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structural insights into ligand recognition and selectivity of the human hydroxycarboxylic acid receptor HCAR2.
Cell Discov, 9, 2023
|
|
8IJD
 
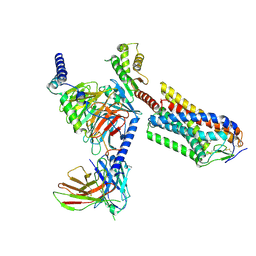 | | Cryo-EM structure of human HCAR2-Gi complex with MK-6892 | | Descriptor: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Pan, X, Fang, Y. | | Deposit date: | 2023-02-27 | | Release date: | 2024-01-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural insights into ligand recognition and selectivity of the human hydroxycarboxylic acid receptor HCAR2.
Cell Discov, 9, 2023
|
|
8IJ3
 
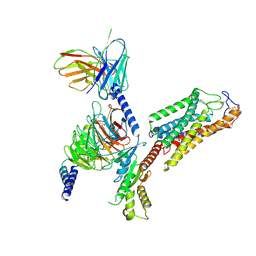 | | Cryo-EM structure of human HCAR2-Gi complex without ligand (apo state) | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Pan, X, Fang, Y. | | Deposit date: | 2023-02-24 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural insights into ligand recognition and selectivity of the human hydroxycarboxylic acid receptor HCAR2.
Cell Discov, 9, 2023
|
|
8IJB
 
 | | Cryo-EM structure of human HCAR2-Gi complex with acipimox | | Descriptor: | 5-methyl-4-oxidanyl-pyrazin-4-ium-2-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Pan, X, Fang, Y. | | Deposit date: | 2023-02-27 | | Release date: | 2024-01-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural insights into ligand recognition and selectivity of the human hydroxycarboxylic acid receptor HCAR2.
Cell Discov, 9, 2023
|
|
8HZM
 
 | | A new fluorescent RNA aptamer bound with N, manganese soak | | Descriptor: | (5~{Z})-5-[[4-[2-hydroxyethyl(methyl)amino]phenyl]methylidene]-3-methyl-2-[(~{E})-2-phenylethenyl]imidazol-4-one, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2023-01-09 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of a small monomeric Clivia fluorogenic RNA with a large Stokes shift.
Nat.Chem.Biol., 2024
|
|
8HZE
 
 | | A new fluorescent RNA aptamer bound with N | | Descriptor: | (5~{Z})-5-[[4-[2-hydroxyethyl(methyl)amino]phenyl]methylidene]-3-methyl-2-[(~{E})-2-phenylethenyl]imidazol-4-one, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2023-01-09 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural basis of a small monomeric Clivia fluorogenic RNA with a large Stokes shift.
Nat.Chem.Biol., 2024
|
|
8HZD
 
 | | A new fluorescent RNA aptamer bound with N618 | | Descriptor: | 4-[(~{E})-2-[(4~{Z})-4-[[3,5-bis(fluoranyl)-4-oxidanyl-phenyl]methylidene]-1-methyl-5-oxidanylidene-imidazol-2-yl]ethenyl]benzenecarbonitrile, MAGNESIUM ION, RNA (36-MER) | | Authors: | Song, Q.Q, Ren, A.M. | | Deposit date: | 2023-01-09 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural basis of a small monomeric Clivia fluorogenic RNA with a large Stokes shift.
Nat.Chem.Biol., 2024
|
|
8HZJ
 
 | | A new fluorescent RNA aptamer bound with N571 | | Descriptor: | (5~{Z})-5-[[3,5-bis(fluoranyl)-4-oxidanyl-phenyl]methylidene]-3-methyl-2-[(~{E})-2-phenylethenyl]imidazol-4-one, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2023-01-09 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of a small monomeric Clivia fluorogenic RNA with a large Stokes shift.
Nat.Chem.Biol., 2024
|
|
