3O07
 
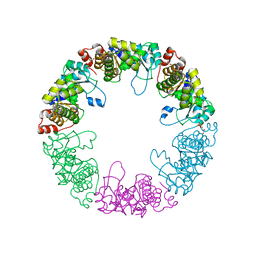 | | Crystal structure of yeast pyridoxal 5-phosphate synthase Snz1 complexed with substrate G3P | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE, Pyridoxine biosynthesis protein SNZ1 | | Authors: | Teng, Y.B, Zhang, X, Hu, H.X, Zhou, C.Z. | | Deposit date: | 2010-07-19 | | Release date: | 2010-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the catalytic mechanism of the yeast pyridoxal 5-phosphate synthase Snz1
Biochem.J., 432, 2010
|
|
7VH2
 
 | |
7VGQ
 
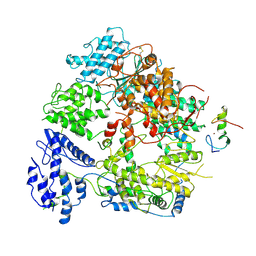 | | Cryo-EM structure of Machupo virus polymerase L in complex with matrix protein Z | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,RING finger protein Z, RNA-directed RNA polymerase L, ZINC ION | | Authors: | Zhang, X, Ma, J, Zhang, S. | | Deposit date: | 2021-09-18 | | Release date: | 2021-09-29 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of Machupo virus polymerase in complex with matrix protein Z.
Nat Commun, 12, 2021
|
|
7VH1
 
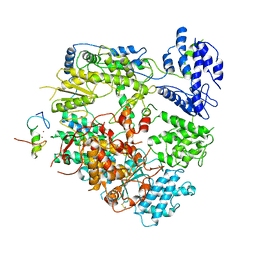 | | Cryo-EM structure of Machupo virus dimeric L-Z complex | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,RING finger protein Z, RNA-directed RNA polymerase L, ZINC ION | | Authors: | Zhang, X, Ma, J, Zhang, S. | | Deposit date: | 2021-09-20 | | Release date: | 2021-09-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of Machupo virus polymerase in complex with matrix protein Z.
Nat Commun, 12, 2021
|
|
5YC0
 
 | | Crystal structure of LP-46/N44 | | Descriptor: | Envelope glycoprotein, LP-46 | | Authors: | Zhang, X, Wang, X, He, Y. | | Deposit date: | 2017-09-05 | | Release date: | 2018-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exceptional potency and structural basis of a T1249-derived lipopeptide fusion inhibitor against HIV-1, HIV-2, and simian immunodeficiency virus
J. Biol. Chem., 293, 2018
|
|
4N6F
 
 | | Crystal structure of Amycolatopsis orientalis BexX complexed with G6P | | Descriptor: | CALCIUM ION, FRUCTOSE -6-PHOSPHATE, Putative thiosugar synthase | | Authors: | Zhang, X, Zhang, Y, Kinsland, C, Sasaki, E, Sun, H.G, Lu, M.J, Liu, T, Ou, A, Li, J, Chen, Y, Liu, H, Ealick, S.E. | | Deposit date: | 2013-10-11 | | Release date: | 2014-05-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Co-opting sulphur-carrier proteins from primary metabolic pathways for 2-thiosugar biosynthesis.
Nature, 509, 2014
|
|
4N6E
 
 | | Crystal structure of Amycolatopsis orientalis BexX/CysO complex | | Descriptor: | Putative thiosugar synthase, SULFATE ION, ThiS/MoaD family protein | | Authors: | Zhang, X, Zhang, Y, Kinsland, C, Sasaki, E, Sun, H.G, Lu, M.J, Liu, T, Ou, A, Li, J, Chen, Y, Liu, H, Ealick, S.E. | | Deposit date: | 2013-10-11 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Co-opting sulphur-carrier proteins from primary metabolic pathways for 2-thiosugar biosynthesis.
Nature, 509, 2014
|
|
5ZII
 
 | |
3RYT
 
 | | The Plexin A1 intracellular region in complex with Rac1 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Plexin-A1, ... | | Authors: | Zhang, X, He, H. | | Deposit date: | 2011-05-11 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.582 Å) | | Cite: | Plexins Are GTPase-Activating Proteins for Rap and Are Activated by Induced Dimerization.
Sci.Signal., 5, 2012
|
|
6IMM
 
 | | Cryo-EM structure of an alphavirus, Sindbis virus | | Descriptor: | Assembly protein E3, Octadecane, Spike glycoprotein E1, ... | | Authors: | Zhang, X, Ma, J, Chen, L. | | Deposit date: | 2018-10-23 | | Release date: | 2019-03-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Implication for alphavirus host-cell entry and assembly indicated by a 3.5 angstrom resolution cryo-EM structure.
Nat Commun, 9, 2018
|
|
4RSQ
 
 | | 2.9A resolution structure of SRPN2 (K198C/E359C) from Anopheles gambiae | | Descriptor: | Serpin 2 | | Authors: | Lovell, S, Battaile, K.P, Zhang, X, Meekins, D.A, An, C, Michel, K. | | Deposit date: | 2014-11-10 | | Release date: | 2014-12-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and Inhibitory Effects of Hinge Loop Mutagenesis in Serpin-2 from the Malaria Vector Anopheles gambiae.
J.Biol.Chem., 290, 2015
|
|
4ROA
 
 | | 1.90A resolution structure of SRPN2 (S358W) from Anopheles gambiae | | Descriptor: | Serpin 2 | | Authors: | Lovell, S, Battaile, K.P, Zhang, X, Meekins, D.A, An, C, Michel, K. | | Deposit date: | 2014-10-28 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Inhibitory Effects of Hinge Loop Mutagenesis in Serpin-2 from the Malaria Vector Anopheles gambiae.
J.Biol.Chem., 290, 2015
|
|
5XWY
 
 | | Electron cryo-microscopy structure of LbuCas13a-crRNA binary complex | | Descriptor: | A type VI-A CRISPR-Cas RNA-guided RNA ribonuclease, Cas13a, RNA (59-MER) | | Authors: | Zhang, X, Wang, Y, Ma, J, Liu, L, Li, X, Li, Z, You, L, Wang, J, Wang, M. | | Deposit date: | 2017-06-30 | | Release date: | 2017-09-13 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The Molecular Architecture for RNA-Guided RNA Cleavage by Cas13a.
Cell, 170, 2017
|
|
4RO9
 
 | | 2.0A resolution structure of SRPN2 (S358E) from Anopheles gambiae | | Descriptor: | GLYCEROL, Serpin 2 | | Authors: | Lovell, S, Battaile, K.P, Zhang, X, Meekins, D.A, An, C, Michel, K. | | Deposit date: | 2014-10-28 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Inhibitory Effects of Hinge Loop Mutagenesis in Serpin-2 from the Malaria Vector Anopheles gambiae.
J.Biol.Chem., 290, 2015
|
|
3T9N
 
 | | Crystal structure of a membrane protein | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Small-conductance mechanosensitive channel | | Authors: | Yang, M, Zhang, X, Ge, J, Wang, J. | | Deposit date: | 2011-08-03 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.456 Å) | | Cite: | Structure and molecular mechanism of an anion-selective mechanosensitive channel of small conductance
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4ROB
 
 | | 2.8A resolution structure of SRPN2 (K198C) from Anopheles gambiae | | Descriptor: | Serpin 2 | | Authors: | Lovell, S, Battaile, K.P, Zhang, X, Meekins, D.A, An, C, Michel, K. | | Deposit date: | 2014-10-28 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 2.8A resolution structure of SRPN2 (K198C) from Anopheles gambiae
To be Published
|
|
5XUR
 
 | | Crystal Structure of Rv2466c C22S Mutant | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Thioredoxin-like reductase Rv2466c | | Authors: | Zhang, X, Li, H. | | Deposit date: | 2017-06-25 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Identification of a Mycothiol-Dependent Nitroreductase from Mycobacterium tuberculosis.
ACS Infect Dis, 4, 2018
|
|
5Y9W
 
 | | Crystal 1 for AtLURE1.2-AtPRK6LRR | | Descriptor: | Pollen receptor-like kinase 6, Protein LURE 1.2, SULFATE ION | | Authors: | Chai, J, Zhang, X. | | Deposit date: | 2017-08-28 | | Release date: | 2017-12-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Structural basis for receptor recognition of pollen tube attraction peptides.
Nat Commun, 8, 2017
|
|
5YAH
 
 | | Crystal 2 for AtLURE1.2-AtPRK6LRR | | Descriptor: | Pollen receptor-like kinase 6, Protein LURE 1.2 | | Authors: | Chai, J, Zhang, X. | | Deposit date: | 2017-08-31 | | Release date: | 2018-04-11 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Structural basis for receptor recognition of pollen tube attraction peptides.
Nat Commun, 8, 2017
|
|
7V66
 
 | | Structure of Apoferritin | | Descriptor: | Ferritin heavy chain | | Authors: | Zhang, X, Wu, C, Shi, H. | | Deposit date: | 2021-08-19 | | Release date: | 2022-10-05 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (1.89 Å) | | Cite: | Low-cooling-rate freezing in biomolecular cryo-electron microscopy for recovery of initial frames.
QRB Discov, 2, 2021
|
|
5ZKZ
 
 | |
5ZIW
 
 | |
7YSK
 
 | | Crystal structure of D-Cysteine desulfhydrase from Pectobacterium atrosepticum | | Descriptor: | D-Cysteine desulfhydrase | | Authors: | Zhang, X, Wang, L, Xu, X, Xing, X, Zhou, J. | | Deposit date: | 2022-08-12 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization and structural basis of D-cysteine desulfhydrase from Pectobacterium atrosepticum
Tetrahedron, 2022
|
|
7YSL
 
 | | Crystal structure of D-Cysteine desulfhydrase with a trapped PLP-pyruvate geminal diamine | | Descriptor: | 1,2-ETHANEDIOL, D-Cysteine desulfhydrase, FORMIC ACID | | Authors: | Zhang, X, Wang, L, Xu, X, Xing, X, Zhou, J. | | Deposit date: | 2022-08-12 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Characterization and structural basis of D-cysteine desulfhydrase from Pectobacterium atrosepticum
Tetrahedron, 2022
|
|
7CTH
 
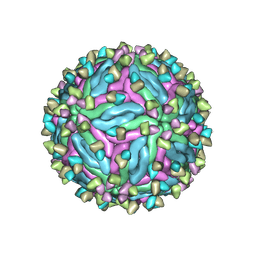 | | Cryo-EM structure of dengue virus serotype 2 in complex with the scFv fragment of the broadly neutralizing antibody EDE1 C10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Core protein, Single Chain Variable Fragment | | Authors: | Zhang, X, Sharma, A, Duquerroy, S, Zhou, Z.H, Rey, F.A. | | Deposit date: | 2020-08-19 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The epitope arrangement on flavivirus particles contributes to Mab C10's extraordinary neutralization breadth across Zika and dengue viruses.
Cell, 184, 2021
|
|
