2DCV
 
 | |
5E13
 
 | | Crystal structure of Eosinophil-derived neurotoxin in complex with the triazole double-headed ribonucleoside 11c | | Descriptor: | 3'-{4-[(4-amino-2-oxopyrimidin-1(2H)-yl)methyl]-1H-1,2,3-triazol-1-yl}-3'-deoxyadenosine, Non-secretory ribonuclease | | Authors: | Chatzileontiadou, D.S.M, Stravodimos, G.A, Kantsadi, A.L, Leonidas, D.D. | | Deposit date: | 2015-09-29 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Triazole double-headed ribonucleosides as inhibitors of eosinophil derived neurotoxin.
Bioorg.Chem., 63, 2015
|
|
8DMB
 
 | | Structure of Desulfovirgula thermocuniculi IsrB (DtIsrB) in complex with omega RNA and target DNA | | Descriptor: | MAGNESIUM ION, Ubiquitin-like protein SMT3,IsrB protein,monomeric superfolder Green Fluorescent Protein, non-target DNA, ... | | Authors: | Seiichi, H, Kappel, K, Zhang, F. | | Deposit date: | 2022-07-08 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the OMEGA nickase IsrB in complex with omega RNA and target DNA.
Nature, 610, 2022
|
|
2ADM
 
 | | ADENINE-N6-DNA-METHYLTRANSFERASE TAQI | | Descriptor: | ADENINE-N6-DNA-METHYLTRANSFERASE TAQI, S-ADENOSYLMETHIONINE | | Authors: | Schluckebier, G, Saenger, W. | | Deposit date: | 1996-07-15 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Differential binding of S-adenosylmethionine S-adenosylhomocysteine and Sinefungin to the adenine-specific DNA methyltransferase M.TaqI.
J.Mol.Biol., 265, 1997
|
|
5KJA
 
 | | Synechocystis apocarotenoid oxygenase (ACO) mutant - Trp149Ala | | Descriptor: | Apocarotenoid-15,15'-oxygenase, CHLORIDE ION, FE (II) ION | | Authors: | Sui, X, Kiser, P.D, Palczewski, K. | | Deposit date: | 2016-06-18 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Key Residues for Catalytic Function and Metal Coordination in a Carotenoid Cleavage Dioxygenase.
J.Biol.Chem., 291, 2016
|
|
2IH5
 
 | |
2IH4
 
 | |
2IH2
 
 | |
3ZK4
 
 | |
2IBS
 
 | | Crystal structure of the adenine-specific DNA methyltransferase M.TaqI complexed with the cofactor analog AETA and a 10 bp DNA containing 2-aminopurine at the target position | | Descriptor: | 5'-D(*GP*AP*CP*AP*TP*CP*GP*(6MA)P*AP*C)-3', 5'-D(*GP*TP*TP*CP*GP*(2PR)P*TP*GP*TP*C)-3', 5'-DEOXY-5'-[2-(AMINO)ETHYLTHIO]ADENOSINE, ... | | Authors: | Pljevaljcic, G, Lenz, T, Scheidig, A.J, Weinhold, E. | | Deposit date: | 2006-09-12 | | Release date: | 2007-05-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2-Aminopurine Flipped into the Active Site of the Adenine-Specific DNA Methyltransferase M.TaqI: Crystal Structures and Time-Resolved Fluorescence
J.Am.Chem.Soc., 129, 2007
|
|
2IBT
 
 | | Crystal structure of the adenine-specific DNA methyltransferase M.TaqI complexed with the cofactor analog AETA and a 10 bp DNA containing 2-aminopurine at the target position and an abasic site analog at the target base partner position | | Descriptor: | 5'-D(*GP*AP*CP*AP*(3DR)P*CP*GP*(6MA)P*AP*C)-3', 5'-D(*GP*TP*TP*CP*GP*(2PR)P*TP*GP*TP*C)-3', 5'-DEOXY-5'-[2-(AMINO)ETHYLTHIO]ADENOSINE, ... | | Authors: | Lenz, T, Scheidig, A.J, Weinhold, E. | | Deposit date: | 2006-09-12 | | Release date: | 2007-05-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 2-Aminopurine Flipped into the Active Site of the Adenine-Specific DNA Methyltransferase M.TaqI: Crystal Structures and Time-Resolved Fluorescence
J.Am.Chem.Soc., 129, 2007
|
|
2NP6
 
 | |
2NP7
 
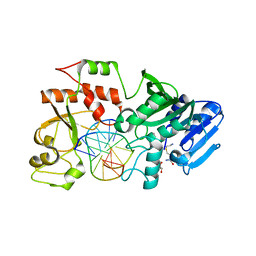 | |
3WS9
 
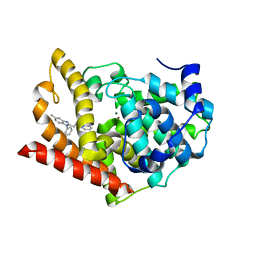 | | Crystal structure of PDE10A in complex with a benzimdazole inhibitor | | Descriptor: | 7-[2-(5-methyl-1-phenyl-1H-benzimidazol-2-yl)ethyl]imidazo[1,5-b]pyridazine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Amano, Y, Honbou, K. | | Deposit date: | 2014-03-04 | | Release date: | 2014-06-04 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Novel benzimidazole derivatives as phosphodiesterase 10A (PDE10A) inhibitors with improved metabolic stability.
Bioorg.Med.Chem., 22, 2014
|
|
3WS8
 
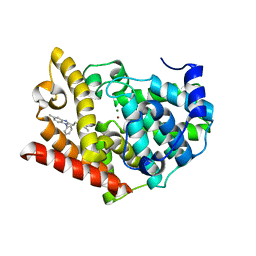 | | Crystal structure of PDE10A in complex with a benzimidazole inhibitor | | Descriptor: | 8-methyl-6-[2-(5-methyl-1-phenyl-1H-benzimidazol-2-yl)ethyl]imidazo[1,5-a]pyrimidine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Amano, Y, Honbou, K. | | Deposit date: | 2014-03-04 | | Release date: | 2014-06-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Novel benzimidazole derivatives as phosphodiesterase 10A (PDE10A) inhibitors with improved metabolic stability.
Bioorg.Med.Chem., 22, 2014
|
|
1GP4
 
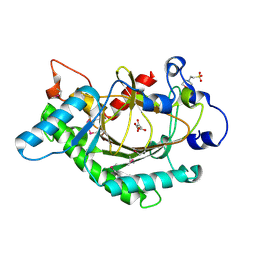 | | Anthocyanidin synthase from Arabidopsis thaliana (selenomethionine substituted) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-OXOGLUTARIC ACID, ANTHOCYANIDIN SYNTHASE | | Authors: | Wilmouth, R.C, Turnbull, J.J, Welford, R.W.D, Clifton, I.J, Prescott, A.G, Schofield, C.J. | | Deposit date: | 2001-10-30 | | Release date: | 2002-02-21 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Mechanism of Anthocyanidin Synthase from Arabidopsis Thaliana.
Structure, 10, 2002
|
|
3V7A
 
 | |
1GP6
 
 | | Anthocyanidin synthase from Arabidopsis thaliana complexed with trans-dihydroquercetin (with 30 min exposure to O2) | | Descriptor: | (2S,3S)-2-(3,4-DIHYDROXYPHENYL)-3,5,7-TRIHYDROXY-2,3-DIHYDRO-4H-CHROMEN-4-ONE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3,5,7,3',4'-PENTAHYDROXYFLAVONE, ... | | Authors: | Wilmouth, R.C, Turnbull, J.J, Welford, R.W.D, Clifton, I.J, Prescott, A.G, Schofield, C.J. | | Deposit date: | 2001-10-30 | | Release date: | 2002-02-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and Mechanism of Anthocyanidin Synthase from Arabidopsis Thaliana.
Structure, 10, 2002
|
|
1GP5
 
 | | Anthocyanidin synthase from Arabidopsis thaliana complexed with trans-dihydroquercetin | | Descriptor: | (2R,3R)-2-(3,4-DIHYDROXYPHENYL)-3,5,7-TRIHYDROXY-2,3-DIHYDRO-4H-CHROMEN-4-ONE, (2S,3S)-2-(3,4-DIHYDROXYPHENYL)-3,5,7-TRIHYDROXY-2,3-DIHYDRO-4H-CHROMEN-4-ONE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Wilmouth, R.C, Turnbull, J.J, Welford, R.W.D, Clifton, I.J, Prescott, A.G, Schofield, C.J. | | Deposit date: | 2001-10-30 | | Release date: | 2002-02-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Mechanism of Anthocyanidin Synthase from Arabidopsis Thaliana.
Structure, 10, 2002
|
|
5GKR
 
 | | Crystal structure of SLE patient-derived anti-DNA antibody in complex with oligonucleotide | | Descriptor: | DNA (5'-D(P*TP*TP*TP*T)-3'), IgG2, Fab (heavy chain), ... | | Authors: | Arimori, T, Sakakibara, S, Kikutani, H, Takagi, J. | | Deposit date: | 2016-07-05 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Clonal evolution and antigen recognition of anti-nuclear antibodies in acute systemic lupus erythematosus
Sci Rep, 7, 2017
|
|
5GKS
 
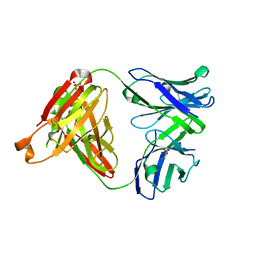 | | Crystal structure of SLE patient-derived anti-DNA antibody | | Descriptor: | IgG2, Fab (heavy chain), PHOSPHATE ION, ... | | Authors: | Arimori, T, Sakakibara, S, Kikutani, H, Takagi, J. | | Deposit date: | 2016-07-05 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Clonal evolution and antigen recognition of anti-nuclear antibodies in acute systemic lupus erythematosus
Sci Rep, 7, 2017
|
|
2I35
 
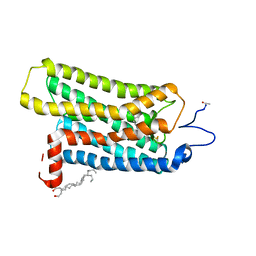 | | Crystal structure of rhombohedral crystal form of ground-state rhodopsin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITIC ACID, RETINAL, ... | | Authors: | Stenkamp, R.E, Le Trong, I, Lodowski, D.T, Salom, D, Palczewski, K. | | Deposit date: | 2006-08-17 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal structure of a photoactivated deprotonated intermediate of rhodopsin.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2I36
 
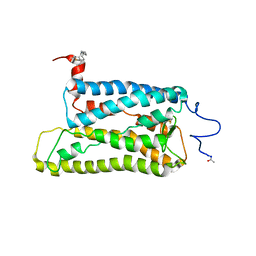 | | Crystal structure of trigonal crystal form of ground-state rhodopsin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITIC ACID, Rhodopsin, ... | | Authors: | Stenkamp, R.E, Le Trong, I, Lodowski, D.T, Salom, D, Palczewski, K. | | Deposit date: | 2006-08-17 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Crystal structure of a photoactivated deprotonated intermediate of rhodopsin.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2I37
 
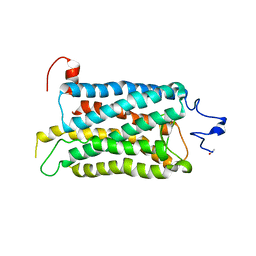 | | Crystal structure of a photoactivated rhodopsin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-beta-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Rhodopsin, ... | | Authors: | Lodowski, D.T, Stenkamp, R.E, Salom, D, Le Trong, I, Palczewski, K. | | Deposit date: | 2006-08-17 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4.15 Å) | | Cite: | Crystal structure of a photoactivated deprotonated intermediate of rhodopsin.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3FSN
 
 | | Crystal structure of RPE65 at 2.14 angstrom resolution | | Descriptor: | FE (II) ION, Retinal pigment epithelium-specific 65 kDa protein, TETRAETHYLENE GLYCOL | | Authors: | Kiser, P.D, Lodowski, D.T, Palczewski, K. | | Deposit date: | 2009-01-11 | | Release date: | 2009-09-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of native RPE65, the retinoid isomerase of the visual cycle.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
