8XA1
 
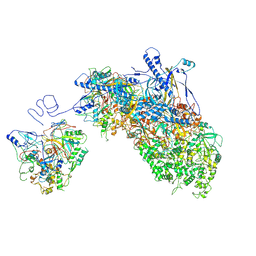 | | Portal vertex capsomer of VZV B-capsid | | Descriptor: | Major capsid protein, Tri1, Tri2A, ... | | Authors: | Nan, W, Lei, C, Xiangxi, W. | | Deposit date: | 2023-12-01 | | Release date: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Insights into varicella-zoster virus assembly from the B- and C-capsid at near-atomic resolution structures
Hlife, 2, 2024
|
|
8X9W
 
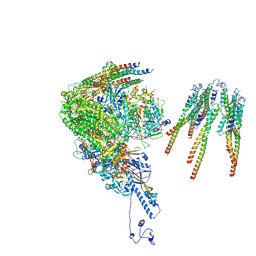 | | portal vertex capsomer of the VZV C-Capsid | | Descriptor: | CVC1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | Authors: | Nan, W, Lei, C, Jiangxi, W. | | Deposit date: | 2023-12-01 | | Release date: | 2025-04-30 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Insights into varicella-zoster virus assembly from the B- and C-capsid at near-atomic resolution structures
Hlife, 2, 2024
|
|
8X9X
 
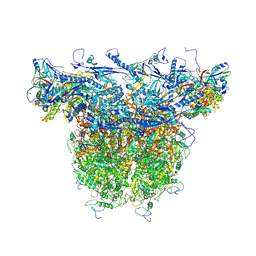 | | C-hexon capsomer of the VZV C-Capsid | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Tri1, ... | | Authors: | Nan, W, Lei, C, Xiangxi, W. | | Deposit date: | 2023-12-01 | | Release date: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Insights into varicella-zoster virus assembly from the B- and C-capsid at near-atomic resolution structures
Hlife, 2, 2024
|
|
8X9Y
 
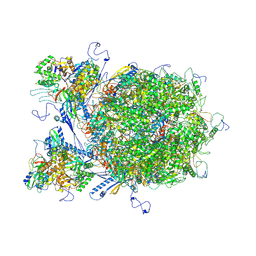 | | E-hexon capsomer of the VZV C-Capsid | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Tri1, ... | | Authors: | Nan, W, Lei, C, Xiangxi, W. | | Deposit date: | 2023-12-01 | | Release date: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Insights into varicella-zoster virus assembly from the B- and C-capsid at near-atomic resolution structures
Hlife, 2, 2024
|
|
7RWF
 
 | | Crystal structure of CDK2 in complex with TW8672 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[2-(1H-indol-3-yl)ethyl]amino}-5-nitrobenzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-08-19 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Development of allosteric and selective CDK2 inhibitors for contraception with negative cooperativity to cyclin binding.
Nat Commun, 14, 2023
|
|
7S84
 
 | | Crystal structure of CDK2 liganded with compound TW8972 | | Descriptor: | 2-{[2-(1H-indol-3-yl)ethyl]amino}-5-(trifluoromethyl)benzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-09-17 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of allosteric and selective CDK2 inhibitors for contraception with negative cooperativity to cyclin binding.
Nat Commun, 14, 2023
|
|
6V51
 
 | | Spin-labeled T4 Lysozyme (9/131FnbY)-(4-Amino-TEMPO) | | Descriptor: | 4-amino-2,2,6,6-tetramethylpiperidin-1-ol, Endolysin | | Authors: | Liu, J, Morizumi, T, Ou, W.L, Wang, L, Ernst, O.P. | | Deposit date: | 2019-12-02 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Genetically Encoded Quinone Methides Enabling Rapid, Site-Specific, and Photocontrolled Protein Modification with Amine Reagents.
J.Am.Chem.Soc., 142, 2020
|
|
8FOW
 
 | | Ternary complex of CDK2 with small molecule ligands TW8672 and Dinaciclib | | Descriptor: | 1,2-ETHANEDIOL, 2-{[2-(1H-indol-3-yl)ethyl]amino}-5-nitrobenzoic acid, 3-[({3-ethyl-5-[(2S)-2-(2-hydroxyethyl)piperidin-1-yl]pyrazolo[1,5-a]pyrimidin-7-yl}amino)methyl]-1-hydroxypyridinium, ... | | Authors: | Schonbrunn, E, Sun, L. | | Deposit date: | 2023-01-03 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Development of allosteric and selective CDK2 inhibitors for contraception with negative cooperativity to cyclin binding.
Nat Commun, 14, 2023
|
|
8FP5
 
 | | CDK2 liganded with ATP and Mg2+ | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Cyclin-dependent kinase 2, ... | | Authors: | Schonbrunn, E, Sun, L. | | Deposit date: | 2023-01-04 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Development of allosteric and selective CDK2 inhibitors for contraception with negative cooperativity to cyclin binding.
Nat Commun, 14, 2023
|
|
8FP0
 
 | |
7Y78
 
 | | Crystal structure of Cry78Aa | | Descriptor: | 1,2-ETHANEDIOL, AMMONIUM ION, Toxin | | Authors: | Cao, B.B, Nie, Y.F, Wang, N.C, Guan, Z.Y, Zhang, D.L, Zhang, J. | | Deposit date: | 2022-06-21 | | Release date: | 2022-08-31 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of Cry78Aa from Bacillus thuringiensis provides insights into its insecticidal activity.
Commun Biol, 5, 2022
|
|
7Y79
 
 | | Crystal structure of Cry78Aa | | Descriptor: | Toxin | | Authors: | Cao, B.B, Nie, Y.F, Wang, N.C, Guan, Z.Y, Zhang, D.L, Zhang, J. | | Deposit date: | 2022-06-21 | | Release date: | 2022-08-31 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | The crystal structure of Cry78Aa from Bacillus thuringiensis provides insights into its insecticidal activity.
Commun Biol, 5, 2022
|
|
7XZ9
 
 | |
7XZD
 
 | |
7XZF
 
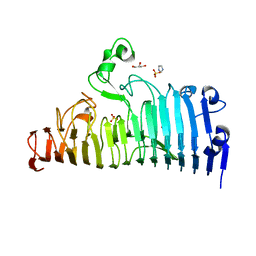 | | Wild type of the N-terminal domain of fucoidan lyase FdlA | | Descriptor: | Fucoidan lyase, GLYCEROL, IMIDAZOLE, ... | | Authors: | Wang, J, Li, M, Pan, X. | | Deposit date: | 2022-06-02 | | Release date: | 2022-09-28 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural and Biochemical Analysis Reveals Catalytic Mechanism of Fucoidan Lyase from Flavobacterium sp. SA-0082.
Mar Drugs, 20, 2022
|
|
7XZ8
 
 | |
7XZB
 
 | |
7XZA
 
 | |
7XZE
 
 | |
7DX4
 
 | | The structure of FC08 Fab-hA.CE2-RBD complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Heavy chain of FC08 Fab, ... | | Authors: | Cao, L, Wang, X. | | Deposit date: | 2021-01-18 | | Release date: | 2021-04-21 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A proof of concept for neutralizing antibody-guided vaccine design against SARS-CoV-2.
Natl Sci Rev, 8, 2021
|
|
7D4G
 
 | |
6KJ9
 
 | | E. coli ATCase catalytic subunit mutant - G128/130A | | Descriptor: | Aspartate carbamoyltransferase catalytic subunit | | Authors: | Lei, Z, Zheng, J, Jia, Z.C. | | Deposit date: | 2019-07-22 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | New regulatory mechanism-based inhibitors of aspartate transcarbamoylase for potential anticancer drug development.
Febs J., 287, 2020
|
|
6KJA
 
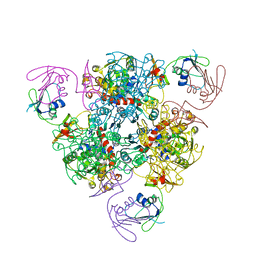 | | E. coli ATCase holoenzyme mutant - G128/130A (catalytic chain) | | Descriptor: | Aspartate carbamoyltransferase catalytic subunit, Aspartate carbamoyltransferase regulatory chain, ZINC ION | | Authors: | Lei, Z, Zheng, J, Jia, Z.C. | | Deposit date: | 2019-07-22 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.064 Å) | | Cite: | New regulatory mechanism-based inhibitors of aspartate transcarbamoylase for potential anticancer drug development.
Febs J., 287, 2020
|
|
6KJ8
 
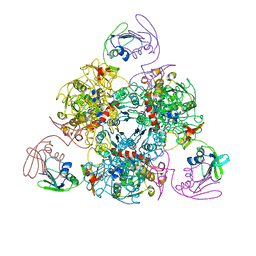 | | E. coli ATCase holoenzyme mutant - G166P (catalytic chain) | | Descriptor: | Aspartate carbamoyltransferase catalytic subunit, Aspartate carbamoyltransferase regulatory chain, ZINC ION | | Authors: | Lei, Z, Zheng, J, Jia, Z.C. | | Deposit date: | 2019-07-22 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.011 Å) | | Cite: | New regulatory mechanism-based inhibitors of aspartate transcarbamoylase for potential anticancer drug development.
Febs J., 287, 2020
|
|
6KJ7
 
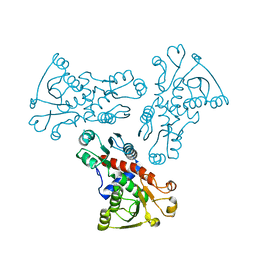 | | E. coli ATCase catalytic subunit mutant - G166P | | Descriptor: | Aspartate carbamoyltransferase catalytic subunit | | Authors: | Lei, Z, Zheng, J, Jia, Z. | | Deposit date: | 2019-07-21 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.839 Å) | | Cite: | New regulatory mechanism-based inhibitors of aspartate transcarbamoylase for potential anticancer drug development.
Febs J., 287, 2020
|
|
