7WO3
 
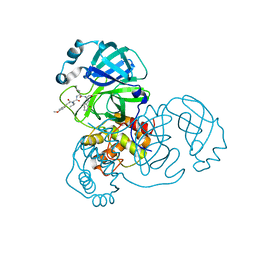 | | SARS-CoV-2 3CLpro | | Descriptor: | (2S)-2-[[(2S)-2-[[(E)-3-(4-methoxyphenyl)prop-2-enoyl]amino]-3-methyl-butanoyl]amino]-4-methyl-N-[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepiperidin-3-yl]propan-2-yl]pentanamide, 3C-like proteinase | | Authors: | Wang, Y, Ye, S. | | Deposit date: | 2022-01-20 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Discovery of SARS-CoV-2 3CL Pro Peptidomimetic Inhibitors through the Catalytic Dyad Histidine-Specific Protein-Ligand Interactions.
Int J Mol Sci, 23, 2022
|
|
7WOF
 
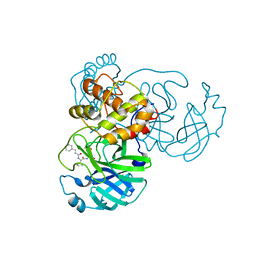 | | SARS-CoV-2 3CLpro | | Descriptor: | (2S,3S)-3-methyl-N-[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepiperidin-3-yl]propan-2-yl]-2-[[(E)-3-phenylprop-2-enoyl]amino]pentanamide, 3C-like proteinase | | Authors: | Wang, Y, Ye, S. | | Deposit date: | 2022-01-21 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of SARS-CoV-2 3CL Pro Peptidomimetic Inhibitors through the Catalytic Dyad Histidine-Specific Protein-Ligand Interactions.
Int J Mol Sci, 23, 2022
|
|
7WO2
 
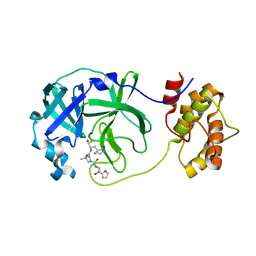 | | SARS-CoV-2 3CLPro Peptidomimetic Inhibitor TPM5 | | Descriptor: | 3C-like proteinase, N-[(2S)-3-methyl-1-[[(2S)-4-methyl-1-oxidanylidene-1-[[(2S)-1-oxidanylidene-3-[(3S}-2-oxidanylidenepiperidin-3-yl]propan-2-yl]amino]pentan-2-yl]amino]-1-oxidanylidene-butan-2-yl]furan-2-carboxamide | | Authors: | Wang, Y, Ye, S. | | Deposit date: | 2022-01-20 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Discovery of SARS-CoV-2 3CL Pro Peptidomimetic Inhibitors through the Catalytic Dyad Histidine-Specific Protein-Ligand Interactions.
Int J Mol Sci, 23, 2022
|
|
6JL0
 
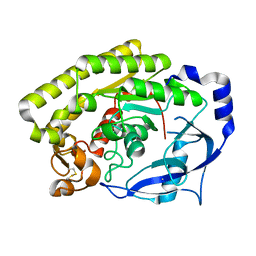 | | Crystal structure of VvPlpA from Vibrio vulnificus | | Descriptor: | BROMIDE ION, Thermolabile hemolysin | | Authors: | Ma, Q, Wan, Y, Liu, C. | | Deposit date: | 2019-03-03 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.073 Å) | | Cite: | Structural analysis of aVibriophospholipase reveals an unusual Ser-His-chloride catalytic triad.
J.Biol.Chem., 294, 2019
|
|
6JL2
 
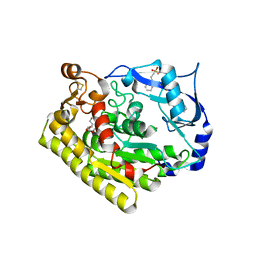 | | Crystal structure of VvPlpA G389N from Vibrio vulnificus | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, HEXAETHYLENE GLYCOL, Thermolabile hemolysin | | Authors: | Ma, Q, Wan, Y, Liu, C. | | Deposit date: | 2019-03-03 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of aVibriophospholipase reveals an unusual Ser-His-chloride catalytic triad.
J.Biol.Chem., 294, 2019
|
|
6JL1
 
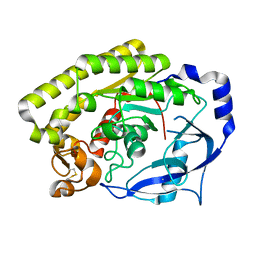 | |
6JKZ
 
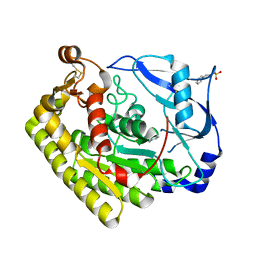 | | Crystal structure of VvPlpA from Vibrio vulnificus | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, CHLORIDE ION, Thermolabile hemolysin | | Authors: | Ma, Q, Wan, Y, Liu, C. | | Deposit date: | 2019-03-03 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.397 Å) | | Cite: | Structural analysis of aVibriophospholipase reveals an unusual Ser-His-chloride catalytic triad.
J.Biol.Chem., 294, 2019
|
|
5WQJ
 
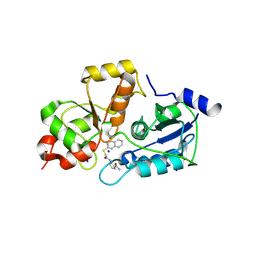 | |
5WQK
 
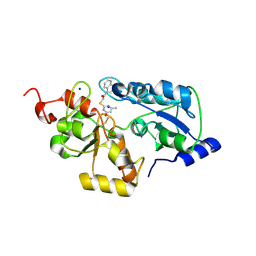 | |
5IY4
 
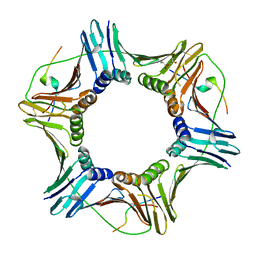 | |
3NYL
 
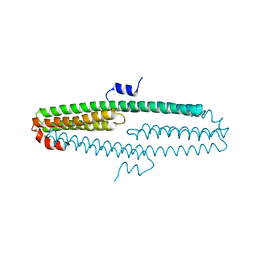 | | The X-ray structure of an antiparallel dimer of the human amyloid precursor protein E2 domain | | Descriptor: | Amyloid beta (A4) protein (Peptidase nexin-II, Alzheimer disease), isoform CRA_b | | Authors: | Ha, Y, Hu, J, Lee, S, Liu, X, Wang, Y. | | Deposit date: | 2010-07-15 | | Release date: | 2011-07-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The X-ray structure of an antiparallel dimer of the human amyloid precursor protein E2 domain.
Mol.Cell, 15, 2004
|
|
5ZED
 
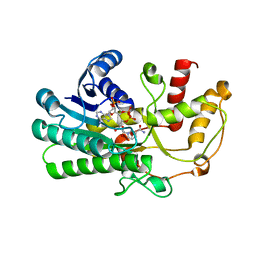 | | Crystal structure of Kluyveromyces polyspora ADH (KpADH) mutant (E214V/T215S) | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Uncharacterized protein ADH | | Authors: | Wang, Y, Zhou, J.Y, Hou, X.D, Xu, G.C, Wu, L, Rao, Y.J, ZHou, J.H, Ni, Y. | | Deposit date: | 2018-02-27 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Structural Insight into Enantioselective Inversion of an Alcohol Dehydrogenase Reveals a "Polar Gate" in Stereorecognition of Diaryl Ketones.
J. Am. Chem. Soc., 140, 2018
|
|
5Z2X
 
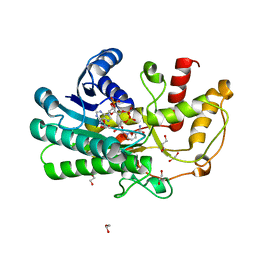 | | Structure of Alcohol dehydrogenase from Kluyveromyces polyspora(KpADH) | | Descriptor: | 1,2-ETHANEDIOL, Alcohol dehydrogenase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wang, Y, Zhou, J.Y, Hou, X.D, Xu, G.C, Wu, L, Rao, Y.J, ZHou, J.H, Ni, Y. | | Deposit date: | 2018-01-04 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Insight into Enantioselective Inversion of an Alcohol Dehydrogenase Reveals a "Polar Gate" in Stereorecognition of Diaryl Ketones.
J. Am. Chem. Soc., 140, 2018
|
|
5ZEC
 
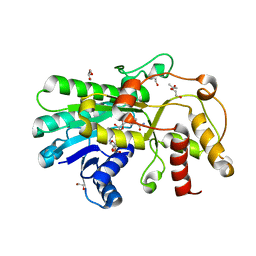 | | Crystal structure of Kluyveromyces polyspora ADH (KpADH) mutant (Q136N/F161V/S196G/E214G/S237C) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ETHANOL, ... | | Authors: | Wang, Y, ZHou, J.Y, Hou, X.D, Xu, G.C, Rao, Y.J, Wu, L, Zhou, J.H, Ni, Y. | | Deposit date: | 2018-02-27 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.779 Å) | | Cite: | Structural Insight into Enantioselective Inversion of an Alcohol Dehydrogenase Reveals a "Polar Gate" in Stereorecognition of Diaryl Ketones.
J. Am. Chem. Soc., 140, 2018
|
|
5XG8
 
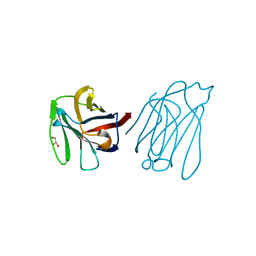 | | Galectin-13/Placental Protein 13 variant R53H crystal structure | | Descriptor: | GLYCEROL, Galactoside-binding soluble lectin 13 | | Authors: | Wang, Y, Su, J.Y. | | Deposit date: | 2017-04-12 | | Release date: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Galectin-13, a different prototype galectin, does not bind beta-galacto-sides and forms dimers via intermolecular disulfide bridges between Cys-136 and Cys-138
Sci Rep, 8, 2018
|
|
6KG6
 
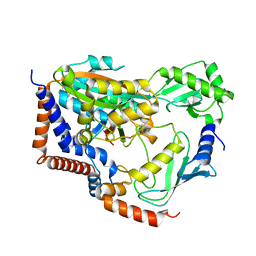 | | Crystal structure of MavC/UBE2N-Ub complex | | Descriptor: | MavC, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-conjugating enzyme E2 N | | Authors: | Wang, Y, Huang, Y, Chang, M, Feng, Y. | | Deposit date: | 2019-07-10 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural insights into the mechanism and inhibition of transglutaminase-induced ubiquitination by the Legionella effector MavC.
Nat Commun, 11, 2020
|
|
6KZD
 
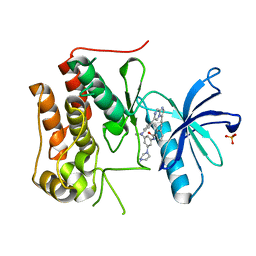 | | Crystal structure of TRKc in complex with 3-((6-(4-aminophenyl)imidazo[1,2-a]pyrazin-3-yl)ethynyl)- N-(3-isopropyl-5-((4-methylpiperazin-1-yl)methyl)phenyl)-2- methylbenzamide | | Descriptor: | 3-[2-[6-(4-aminophenyl)imidazo[1,2-a]pyrazin-3-yl]ethynyl]-2-methyl-~{N}-[3-(4-methylpiperazin-1-yl)-5-propan-2-yl-phenyl]benzamide, NT-3 growth factor receptor, PHOSPHATE ION | | Authors: | Wang, Y, Zhang, Z.M. | | Deposit date: | 2019-09-23 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.708 Å) | | Cite: | Design, synthesis and biological evaluation of 3-(imidazo[1,2-a]pyrazin-3-ylethynyl)-2-methylbenzamides as potent and selective pan-tropomyosin receptor kinase (TRK) inhibitors.
Eur.J.Med.Chem., 179, 2019
|
|
1FB9
 
 | |
8SPP
 
 | |
7BXG
 
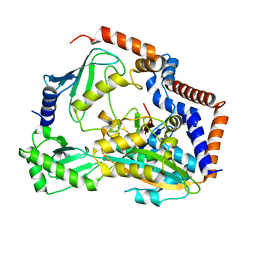 | | MavC-UBE2N-Ub complex | | Descriptor: | MavC, Polyubiquitin-C, Ubiquitin-conjugating enzyme E2 N | | Authors: | Gao, P, Wang, Y. | | Deposit date: | 2020-04-19 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | Insights into catalysis and regulation of non-canonical ubiquitination and deubiquitination by bacterial deamidase effectors.
Nat Commun, 11, 2020
|
|
7BXH
 
 | | MavC-Lpg2149 complex | | Descriptor: | Lpg2149, MavC | | Authors: | Gao, P, Wang, Y. | | Deposit date: | 2020-04-19 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insights into catalysis and regulation of non-canonical ubiquitination and deubiquitination by bacterial deamidase effectors.
Nat Commun, 11, 2020
|
|
5H1K
 
 | | Crystal structure of WD40 repeat domains of Gemin5 in complex with 13-nt U4 snRNA fragment | | Descriptor: | Gem-associated protein 5, U4 snRNA (5'-R(*GP*CP*AP*AP*UP*UP*UP*UP*UP*GP*AP*CP*A)-3') | | Authors: | Wang, Y, Jin, W, Liu, C.P, Yang, N, Jin, M, Cong, Y, Wang, M, Xu, R.M. | | Deposit date: | 2016-10-10 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for snRNA recognition by the double-WD40 repeat domain of Gemin5
Genes Dev., 30, 2016
|
|
2R6W
 
 | | Estrogen receptor alpha ligand-binding domain complexed to a SERM | | Descriptor: | Estrogen receptor, [6-hydroxy-2-(4-hydroxyphenyl)-1-benzothien-3-yl]{4-[2-(4-methylpiperidin-1-yl)ethoxy]phenyl}methanone | | Authors: | Wang, Y. | | Deposit date: | 2007-09-06 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Prediction of the tissue-specificity of selective estrogen receptor modulators by using a single biochemical method.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2R6Y
 
 | | Estrogen receptor alpha ligand-binding domain in complex with a SERM | | Descriptor: | Estrogen receptor, [6-hydroxy-2-(4-hydroxyphenyl)-1-benzothien-3-yl][4-(2-pyrrolidin-1-ylethoxy)phenyl]methanone | | Authors: | Wang, Y. | | Deposit date: | 2007-09-06 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Prediction of the tissue-specificity of selective estrogen receptor modulators by using a single biochemical method.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3ZG6
 
 | | The novel de-long chain fatty acid function of human sirt6 | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, GLYCEROL, NAD-DEPENDENT PROTEIN DEACETYLASE SIRTUIN-6, ... | | Authors: | Wang, Y, Hao, Q. | | Deposit date: | 2012-12-15 | | Release date: | 2013-04-03 | | Last modified: | 2013-04-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Sirt6 Regulates Tnf-Alpha Secretion Through Hydrolysis of Long-Chain Fatty Acyl Lysine
Nature, 496, 2013
|
|
