4Y9J
 
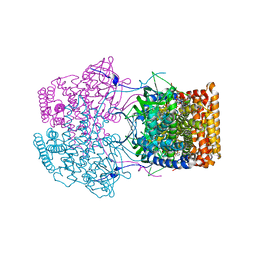 | |
6IEC
 
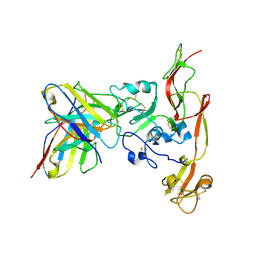 | | Structure of RVFV Gn and human monoclonal antibody R17 | | Descriptor: | NSmGnGc, R17 H chain, R17 L chain | | Authors: | Wang, Q.H, Wu, Y, Gao, F, Qi, J.X, Gao, G.F. | | Deposit date: | 2018-09-13 | | Release date: | 2019-04-10 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Neutralization mechanism of human monoclonal antibodies against Rift Valley fever virus.
Nat Microbiol, 4, 2019
|
|
6IEK
 
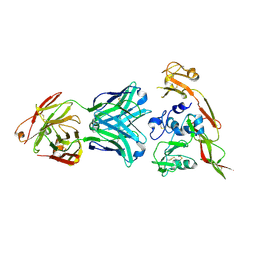 | | Structure of RVFV Gn and human monoclonal antibody R12 | | Descriptor: | Heavy chain of Fab R12, Light chain of Fab R12, NSmGnGc | | Authors: | Wang, Q.H, Wu, Y, Gao, F, Qi, J.X, Gao, G.F. | | Deposit date: | 2018-09-14 | | Release date: | 2019-04-10 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Neutralization mechanism of human monoclonal antibodies against Rift Valley fever virus.
Nat Microbiol, 4, 2019
|
|
8HWT
 
 | |
8HWS
 
 | | The complex structure of Omicron BA.4 RBD with BD604, S309, and S304 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, BD-604 Fab Heavy chain, BD-604 Fab Light chain, ... | | Authors: | He, Q.W, Xu, Z.P, Xie, Y.F. | | Deposit date: | 2023-01-02 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | An updated atlas of antibody evasion by SARS-CoV-2 Omicron sub-variants including BQ.1.1 and XBB.
Cell Rep Med, 4, 2023
|
|
6JO7
 
 | | Crystal structure of mouse MXRA8 | | Descriptor: | Matrix remodeling-associated protein 8 | | Authors: | Song, H, Zhao, Z, Qi, J, Gao, F, Gao, G.F. | | Deposit date: | 2019-03-20 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Basis of Arthritogenic Alphavirus Receptor MXRA8 Binding to Chikungunya Virus Envelope Protein.
Cell, 177, 2019
|
|
6JO8
 
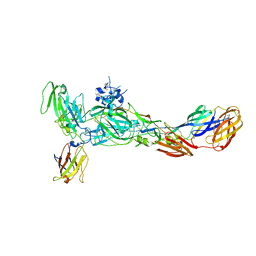 | | The complex structure of CHIKV envelope glycoprotein bound to human MXRA8 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHIKV E1, ... | | Authors: | Song, H, Zhao, Z, Qi, J, Gao, F, Gao, F.G. | | Deposit date: | 2019-03-20 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.495 Å) | | Cite: | Molecular Basis of Arthritogenic Alphavirus Receptor MXRA8 Binding to Chikungunya Virus Envelope Protein.
Cell, 177, 2019
|
|
5XLP
 
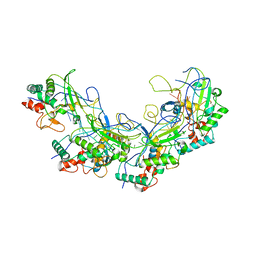 | |
5XHQ
 
 | | Apolipoprotein N-acyl Transferase | | Descriptor: | Apolipoprotein N-acyltransferase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Yingzhi, X, Yong, X, Guangyuan, L, Fei, S. | | Deposit date: | 2017-04-23 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.587 Å) | | Cite: | Crystal structure of E. coli apolipoprotein N-acyl transferase
Nat Commun, 8, 2017
|
|
5XLO
 
 | |
7BZN
 
 | | Cryo-EM structure of mature Coxsackievirus A10 at pH 7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7BZO
 
 | | Cryo-EM structure of mature Coxsackievirus A10 at pH 5.5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7BZU
 
 | | Cryo-EM structure of mature Coxsackievirus A10 in complex with KRM1 at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid protein VP1, Capsid protein VP2, ... | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-22 | | Last modified: | 2020-08-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7BZT
 
 | | Cryo-EM structure of mature Coxsackievirus A10 in complex with KRM1 at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid protein VP1, Capsid protein VP2, ... | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-22 | | Last modified: | 2020-08-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7BSD
 
 | |
7BSC
 
 | | Complex structure of 1G5.3 Fab bound to DENV2 NS1c | | Descriptor: | 1G5.3 Fab Heavy Chain, 1G5.3 Fab Light Chain, Non-structural protein 1 | | Authors: | Song, H, Qi, J, Gao, F.G. | | Deposit date: | 2020-03-30 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A broadly protective antibody that targets the flavivirus NS1 protein.
Science, 371, 2021
|
|
7BPK
 
 | | Zika virus envelope protein mutant bound to mAb | | Descriptor: | Envelope protein, IG c307_light_IGLV1-51_IGLJ2, Z3L1 Heavy chain | | Authors: | Dai, L, Qi, J, Gao, G.F. | | Deposit date: | 2020-03-23 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Protective Zika vaccines engineered to eliminate enhancement of dengue infection via immunodominance switch.
Nat.Immunol., 22, 2021
|
|
7BQ5
 
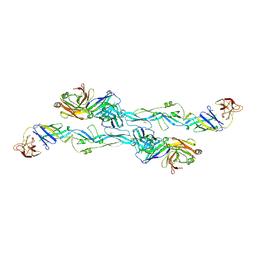 | | ZIKV sE bound to mAb Z6 | | Descriptor: | Z6 Light Chain, Z6 heavy chain, envelope protein | | Authors: | Dai, L, Qi, J, Gao, G.F. | | Deposit date: | 2020-03-24 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Protective Zika vaccines engineered to eliminate enhancement of dengue infection via immunodominance switch.
Nat.Immunol., 22, 2021
|
|
8HPU
 
 | |
8HQ7
 
 | |
8HPF
 
 | |
8HRD
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Delta variant in complex with IMCAS74 Fab and W14 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IMCAS74 Fab heavy chain, IMCAS74 Fab light chain, ... | | Authors: | Zhao, R.C, Wu, L.L, Han, P. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Defining a de novo non-RBM antibody as RBD-8 and its synergistic rescue of immune-evaded antibodies to neutralize Omicron SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8HPV
 
 | |
3J1P
 
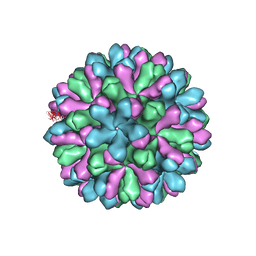 | | Atomic model of rabbit hemorrhagic disease virus | | Descriptor: | Major capsid protein VP60 | | Authors: | Wang, X, Liu, Y, Sun, F. | | Deposit date: | 2012-04-09 | | Release date: | 2013-01-30 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Atomic model of rabbit hemorrhagic disease virus by cryo-electron microscopy and crystallography.
Plos Pathog., 9, 2013
|
|
4EJR
 
 | | Crystal structure of major capsid protein S domain from rabbit hemorrhagic disease virus | | Descriptor: | Major capsid protein VP60 | | Authors: | Xu, F, Ma, J, Zhang, K, Wang, X, Sun, F. | | Deposit date: | 2012-04-07 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Atomic model of rabbit hemorrhagic disease virus by cryo-electron microscopy and crystallography.
Plos Pathog., 9, 2013
|
|
