4TPW
 
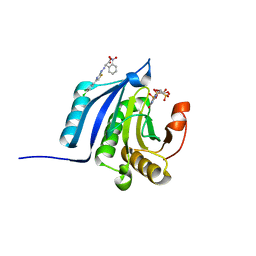 | | The co-complex structure of the translation initiation factor eIF4E with the inhibitor 4EGI-1 reveals an allosteric mechanism for dissociating eIF4G | | Descriptor: | (2E)-2-{2-[4-(3,4-dichlorophenyl)-1,3-thiazol-2-yl]hydrazinylidene}-3-(2-nitrophenyl)propanoic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papadopoulos, E, Jenni, S, Wagner, G. | | Deposit date: | 2014-06-09 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the eukaryotic translation initiation factor eIF4E in complex with 4EGI-1 reveals an allosteric mechanism for dissociating eIF4G.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4TQB
 
 | | The co-complex structure of the translation initiation factor eIF4E with the inhibitor 4EGI-1 reveals an allosteric mechanism for dissociating eIF4G | | Descriptor: | (2E)-2-{2-[4-(4-bromophenyl)-1,3-thiazol-2-yl]hydrazinylidene}-3-(2-nitrophenyl)propanoic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papadopoulos, E, Jenni, S, Wagner, G. | | Deposit date: | 2014-06-10 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure of the eukaryotic translation initiation factor eIF4E in complex with 4EGI-1 reveals an allosteric mechanism for dissociating eIF4G.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5OKP
 
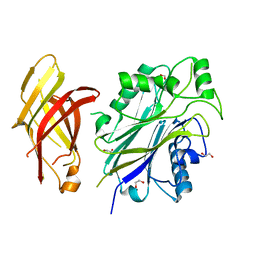 | |
5OKM
 
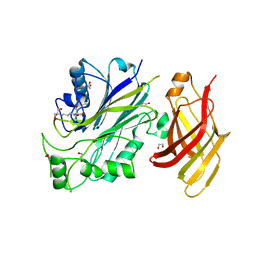 | | Crystal structure of human SHIP2 Phosphatase-C2 | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, NITRATE ION, ... | | Authors: | Le Coq, J, Lietha, D. | | Deposit date: | 2017-07-25 | | Release date: | 2017-08-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural basis for interdomain communication in SHIP2 providing high phosphatase activity.
Elife, 6, 2017
|
|
5OKN
 
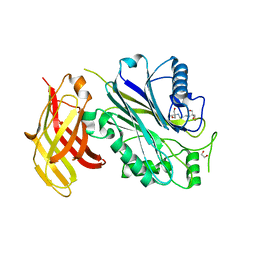 | | Crystal structure of human SHIP2 Phosphatase-C2 D607A mutant | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, ... | | Authors: | Le Coq, J, Lietha, D. | | Deposit date: | 2017-07-25 | | Release date: | 2017-08-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis for interdomain communication in SHIP2 providing high phosphatase activity.
Elife, 6, 2017
|
|
5OKO
 
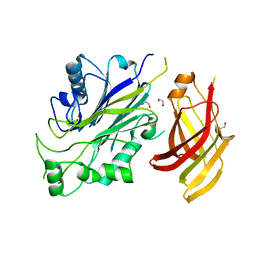 | | Crystal structure of human SHIP2 Phosphatase-C2 double mutant F593D/L597D | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 2 | | Authors: | Le Coq, J, Lietha, D. | | Deposit date: | 2017-07-25 | | Release date: | 2017-08-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural basis for interdomain communication in SHIP2 providing high phosphatase activity.
Elife, 6, 2017
|
|
2PLV
 
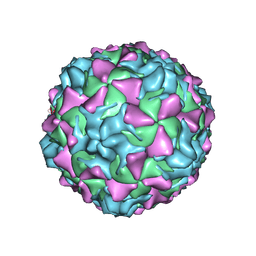 | |
7PFP
 
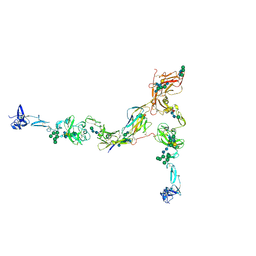 | | Full-length cryo-EM structure of the native human uromodulin (UMOD)/Tamm-Horsfall protein (THP) filament | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Uromodulin, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jovine, L, Xu, C, Stsiapanava, A, Carroni, M, Tunyasuvunakool, K, Jumper, J, Wu, B. | | Deposit date: | 2021-08-11 | | Release date: | 2022-03-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7Q3N
 
 | | Cryo-EM of the complex between human uromodulin (UMOD)/Tamm-Horsfall protein (THP) and the FimH lectin domain from uropathogenic E. coli | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Type 1 fimbiral adhesin FimH, ... | | Authors: | Jovine, L, Xu, C, Stsiapanava, A, Carroni, M, Tunyasuvunakool, K, Jumper, J, Wu, B. | | Deposit date: | 2021-10-28 | | Release date: | 2022-03-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8EOO
 
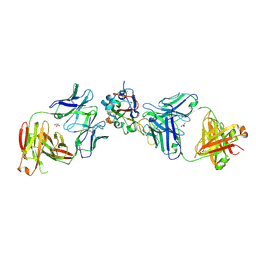 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing human antibodies WRAIR-2063 and WRAIR-2151 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Spike protein S1, ... | | Authors: | Jensen, J.L, Sankhala, R.S, Joyce, M.G. | | Deposit date: | 2022-10-03 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Targeting the Spike Receptor Binding Domain Class V Cryptic Epitope by an Antibody with Pan-Sarbecovirus Activity.
J.Virol., 97, 2023
|
|
8EEE
 
 | |
8EEZ
 
 | |
8EF3
 
 | |
8EE8
 
 | |
8EE5
 
 | |
8EED
 
 | |
8EF0
 
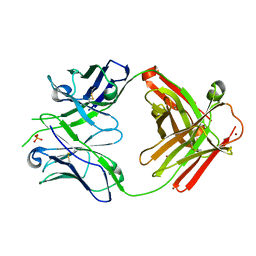 | |
8EF1
 
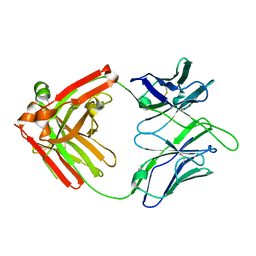 | |
8EF2
 
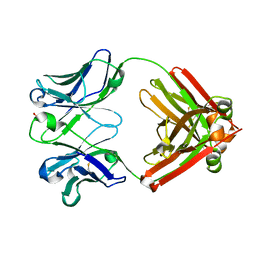 | |
7QGC
 
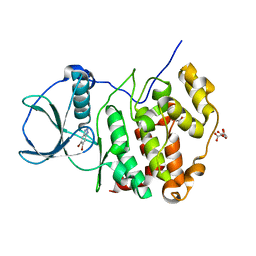 | | H. SAPIENS CK2 KINASE ALPHA SUBUNIT IN COMPLEX WITH THE ATP-COMPETITIVE INHIBITOR 5,6-DIBROMOBENZOTRIAZOLE AT PH 5.5 | | Descriptor: | 5,6-DIBROMOBENZOTRIAZOLE, CITRATE ANION, Casein kinase II subunit alpha, ... | | Authors: | Winiewska-Szajewska, M, Czapinska, H, Kaus-Drobek, M, Piasecka, A, Mieczkowska, K, Dadlez, M, Bochtler, M, Poznanski, J. | | Deposit date: | 2021-12-08 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Competition between electrostatic interactions and halogen bonding in the protein-ligand system: structural and thermodynamic studies of 5,6-dibromobenzotriazole-hCK2 alpha complexes.
Sci Rep, 12, 2022
|
|
7QGE
 
 | | H. SAPIENS CK2 KINASE ALPHA SUBUNIT IN COMPLEX WITH THE ATP-COMPETITIVE INHIBITOR 5,6,7,8-TETRABROMOBENZOTRIAZOLE (TBBt) AT PH 8.5 | | Descriptor: | 4,5,6,7-TETRABROMOBENZOTRIAZOLE, CHLORIDE ION, Casein kinase II subunit alpha, ... | | Authors: | Winiewska-Szajewska, M, Czapinska, H, Kaus-Drobek, M, Piasecka, A, Mieczkowska, K, Dadlez, M, Bochtler, M, Poznanski, J. | | Deposit date: | 2021-12-08 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Competition between electrostatic interactions and halogen bonding in the protein-ligand system: structural and thermodynamic studies of 5,6-dibromobenzotriazole-hCK2 alpha complexes.
Sci Rep, 12, 2022
|
|
7QGD
 
 | | H. SAPIENS CK2 KINASE ALPHA SUBUNIT IN COMPLEX WITH THE ATP-COMPETITIVE INHIBITOR 5,6-DIBROMOBENZOTRIAZOLE AT PH 8.5 | | Descriptor: | 5,6-DIBROMOBENZOTRIAZOLE, CHLORIDE ION, Casein kinase II subunit alpha, ... | | Authors: | Winiewska-Szajewska, M, Czapinska, H, Kaus-Drobek, M, Piasecka, A, Mieczkowska, K, Dadlez, M, Bochtler, M, Poznanski, J. | | Deposit date: | 2021-12-08 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Competition between electrostatic interactions and halogen bonding in the protein-ligand system: structural and thermodynamic studies of 5,6-dibromobenzotriazole-hCK2 alpha complexes.
Sci Rep, 12, 2022
|
|
7QGB
 
 | | H. SAPIENS CK2 KINASE ALPHA SUBUNIT IN COMPLEX WITH THE ATP-COMPETITIVE INHIBITOR 5,6-DIBROMOBENZOTRIAZOLE AT PH 6.5 | | Descriptor: | 5,6-DIBROMOBENZOTRIAZOLE, Casein kinase II subunit alpha | | Authors: | Winiewska-Szajewska, M, Czapinska, H, Kaus-Drobek, M, Piasecka, A, Mieczkowska, K, Dadlez, M, Bochtler, M, Poznanski, J. | | Deposit date: | 2021-12-08 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Competition between electrostatic interactions and halogen bonding in the protein-ligand system: structural and thermodynamic studies of 5,6-dibromobenzotriazole-hCK2 alpha complexes.
Sci Rep, 12, 2022
|
|
1ZTO
 
 | | INACTIVATION GATE OF POTASSIUM CHANNEL RCK4, NMR, 8 STRUCTURES | | Descriptor: | POTASSIUM CHANNEL PROTEIN RCK4 | | Authors: | Antz, C, Geyer, M, Fakler, B, Schott, M, Frank, R, Guy, H.R, Ruppersberg, J.P, Kalbitzer, H.R. | | Deposit date: | 1996-11-15 | | Release date: | 1997-06-05 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR structure of inactivation gates from mammalian voltage-dependent potassium channels.
Nature, 385, 1997
|
|
7P6S
 
 | | Crystal structure of the FimH-binding decoy module of human glycoprotein 2 (GP2) (crystal form II) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform Alpha of Pancreatic secretory granule membrane major glycoprotein GP2, pentane-1,5-diol | | Authors: | Stsiapanava, A, Tunyasuvunakool, K, Jumper, J, de Sanctis, D, Jovine, L. | | Deposit date: | 2021-07-17 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
