3STD
 
 | | SCYTALONE DEHYDRATASE AND CYANOCINNOLINE INHIBITOR | | Descriptor: | 4-(3,3-diphenylpropylamino)cinnoline-3-carbonitrile, CALCIUM ION, PROTEIN (SCYTALONE DEHYDRATASE) | | Authors: | Chen, J.M, Xu, S.L, Wawrzak, Z, Basarab, G.S, Jordan, D.B. | | Deposit date: | 1998-10-16 | | Release date: | 1999-10-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-based design of potent inhibitors of scytalone dehydratase: displacement of a water molecule from the active site.
Biochemistry, 37, 1998
|
|
1A30
 
 | | HIV-1 PROTEASE COMPLEXED WITH A TRIPEPTIDE INHIBITOR | | Descriptor: | HIV-1 PROTEASE, TRIPEPTIDE GLU-ASP-LEU | | Authors: | Louis, J.M, Dyda, F, Nashed, N.T, Kimmel, A.R, Davies, D.R. | | Deposit date: | 1998-01-27 | | Release date: | 1998-04-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hydrophilic peptides derived from the transframe region of Gag-Pol inhibit the HIV-1 protease.
Biochemistry, 37, 1998
|
|
1A8K
 
 | | CRYSTALLOGRAPHIC ANALYSIS OF HUMAN IMMUNODEFICIENCY VIRUS 1 PROTEASE WITH AN ANALOG OF THE CONSERVED CA-P2 SUBSTRATE: INTERACTIONS WITH FREQUENTLY OCCURRING GLUTAMIC ACID RESIDUE AT P2' POSITION OF SUBSTRATES | | Descriptor: | HIV PROTEASE, N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide | | Authors: | Weber, I.T, Wu, J, Adomat, J, Harrison, R.W, Kimmel, A.R, Wondrak, E.M, Louis, J.M. | | Deposit date: | 1998-03-27 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic analysis of human immunodeficiency virus 1 protease with an analog of the conserved CA-p2 substrate -- interactions with frequently occurring glutamic acid residue at P2' position of substrates.
Eur.J.Biochem., 249, 1997
|
|
1BIW
 
 | | DESIGN AND SYNTHESIS OF CONFORMATIONALLY-CONSTRAINED MMP INHIBITORS | | Descriptor: | CALCIUM ION, N1-HYDROXY-2-(3-HYDROXY-PROPYL)-3-ISOBUTYL-N4-[1-(2-METHOXY-ETHYL)-2-OXO-AZEPAN-3-YL]-SUCCINAMIDE, PROTEIN (STROMELYSIN-1 COMPLEX), ... | | Authors: | Natchus, M.G, Cheng, M, Wahl, C.T, Pikul, S, Almstead, N.G, Bradley, R.S, Taiwo, Y.O, Mieling, G.E, Dunaway, C.M, Snider, C.E, McIver, J.M, Barnett, B.L, McPhail, S.J, Anastasio, M.B, De, B. | | Deposit date: | 1998-06-19 | | Release date: | 1999-07-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design and synthesis of conformationally-constrained MMP inhibitors.
Bioorg.Med.Chem.Lett., 8, 1998
|
|
1BWT
 
 | |
1BY8
 
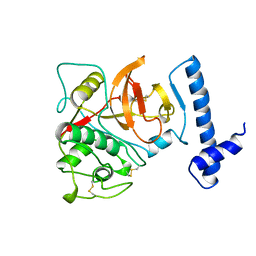 | | THE CRYSTAL STRUCTURE OF HUMAN PROCATHEPSIN K | | Descriptor: | PROTEIN (PROCATHEPSIN K) | | Authors: | Lalonde, J.M, Zhao, B, Smith, W.W, Janson, C.A, Desjarlais, R.L, Tomaszek, T.A, Carr, T.J, Thompson, S.K, Yamashita, D.S, Veber, D.F, Abdel-Mequid, S.S. | | Deposit date: | 1998-10-27 | | Release date: | 1999-10-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of human procathepsin K.
Biochemistry, 38, 1999
|
|
3SN4
 
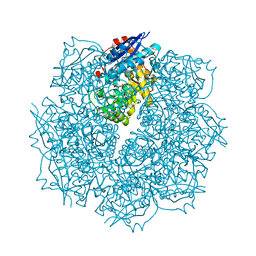 | | Crystal structure of putative L-alanine-DL-glutamate epimerase from Burkholderia xenovorans strain LB400 bound to magnesium and alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Bonanno, J.B, Patskovsky, Y, Toro, R, Dickey, M, Bain, K.T, Wu, B, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-06-28 | | Release date: | 2011-07-27 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of putative L-alanine-DL-glutamate epimerase from Burkholderia xenovorans strain LB400 bound to magnesium and alpha-ketoglutarate
To be Published
|
|
3SA2
 
 | |
3SN1
 
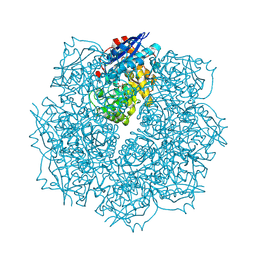 | | Crystal structure of putative L-alanine-DL-glutamate epimerase from Burkholderia xenovorans strain LB400 bound to magnesium and tartrate | | Descriptor: | CHLORIDE ION, D(-)-TARTARIC ACID, MAGNESIUM ION, ... | | Authors: | Bonanno, J.B, Patskovsky, Y, Toro, R, Dickey, M, Bain, K.T, Wu, B, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-06-28 | | Release date: | 2011-07-27 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of putative L-alanine-DL-glutamate epimerase from Burkholderia xenovorans strain LB400 bound to magnesium and tartrate
To be Published
|
|
3S48
 
 | | Human Alpha-Haemoglobin Complexed with the First NEAT Domain of IsdH from Staphylococcus aureus | | Descriptor: | Hemoglobin subunit alpha, Iron-regulated surface determinant protein H, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kumar, K.K, Jacques, D.A, Caradoc-Davies, T.T, Guss, J.M, Gell, D.A. | | Deposit date: | 2011-05-19 | | Release date: | 2012-05-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | The structure of alpha-haemoglobin in complex with a haemoglobin-binding domain from Staphylococcus aureus reveals the elusive alpha-haemoglobin dimerization interface
ACTA CRYSTALLOGR.,SECT.F, 70, 2014
|
|
8JJH
 
 | | Crystal structure of QH-hNTAQ1 C28S | | Descriptor: | Protein N-terminal glutamine amidohydrolase | | Authors: | Kang, J.M, Han, B.W. | | Deposit date: | 2023-05-30 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
1B8I
 
 | | STRUCTURE OF THE HOMEOTIC UBX/EXD/DNA TERNARY COMPLEX | | Descriptor: | DNA (5'-D(*AP*CP*GP*TP*GP*AP*TP*TP*TP*AP*TP*GP*GP*CP*G)-3'), DNA (5'-D(*GP*TP*CP*GP*CP*CP*AP*TP*AP*AP*AP*TP*CP*AP*C)-3'), PROTEIN (HOMEOBOX PROTEIN EXTRADENTICLE), ... | | Authors: | Passner, J.M, Ryoo, H.-D, Shen, L, Mann, R.S, Aggarwal, A.K. | | Deposit date: | 1999-02-01 | | Release date: | 1999-04-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a DNA-bound Ultrabithorax-Extradenticle homeodomain complex.
Nature, 397, 1999
|
|
1B13
 
 | | CLOSTRIDIUM PASTEURIANUM RUBREDOXIN G10A MUTANT | | Descriptor: | FE (III) ION, PROTEIN (RUBREDOXIN) | | Authors: | Maher, M.J, Guss, J.M, Wilce, M.C.J, Wedd, A.G. | | Deposit date: | 1998-11-26 | | Release date: | 1999-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Rubredoxin from Clostridium pasteurianum. Structures of G10A, G43A and G10VG43A mutant proteins. Mutation of conserved glycine 10 to valine causes the 9-10 peptide link to invert.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1B45
 
 | | ALPHA-CNIA CONOTOXIN FROM CONUS CONSORS, NMR, 43 STRUCTURES | | Descriptor: | ALPHA-CNIA | | Authors: | Favreau, P, Krimm, I, Le Gall, F, Bobenrieth, M.J, Lamthanh, H, Bouet, F, Servent, D, Molgo, J, Menez, A, Letourneux, Y, Lancelin, J.M. | | Deposit date: | 1999-01-05 | | Release date: | 1999-07-09 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Biochemical characterization and nuclear magnetic resonance structure of novel alpha-conotoxins isolated from the venom of Conus consors.
Biochemistry, 38, 1999
|
|
1AG7
 
 | | CONOTOXIN GS, NMR, 20 STRUCTURES | | Descriptor: | CONOTOXIN GS | | Authors: | Hill, J.M, Alewood, P.F, Craik, D.J. | | Deposit date: | 1997-04-03 | | Release date: | 1998-04-08 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the sodium channel antagonist conotoxin GS: a new molecular caliper for probing sodium channel geometry.
Structure, 5, 1997
|
|
4JXV
 
 | |
1A16
 
 | | AMINOPEPTIDASE P FROM E. COLI WITH THE INHIBITOR PRO-LEU | | Descriptor: | AMINOPEPTIDASE P, LEUCINE, MANGANESE (II) ION, ... | | Authors: | Wilce, M.C, Bond, C.S, Lilley, P.E, Dixon, N.E, Freeman, H.C, Guss, J.M. | | Deposit date: | 1997-12-22 | | Release date: | 1999-04-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and mechanism of a proline-specific aminopeptidase from Escherichia coli.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
4K2G
 
 | | Structure of Pseudomonas aeruginosa PvdQ bound to BRD-A33442372 | | Descriptor: | (2S)-(4-fluorophenyl)[6-(trifluoromethyl)pyridin-2-yl]ethanenitrile, 1,2-ETHANEDIOL, Acyl-homoserine lactone acylase PvdQ | | Authors: | Drake, E.J, Wurst, J.M, Theriault, J.R, Munoz, B, Gulick, A.M. | | Deposit date: | 2013-04-09 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of Inhibitors of PvdQ, an Enzyme Involved in the Synthesis of the Siderophore Pyoverdine.
Acs Chem.Biol., 9, 2014
|
|
4K1E
 
 | | Atomic resolution crystal structures of Kallikrein-Related Peptidase 4 complexed with a modified SFTI inhibitor FCQR | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Kallikrein-4, LITHIUM ION, ... | | Authors: | Ilyichova, O.V, Swedberg, J.E, de Veer, S.J, Sit, K.C, Harris, J.M, Buckle, A.M. | | Deposit date: | 2013-04-04 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Direct and indirect mechanisms of KLK4 inhibition revealed by structure and dynamics
Sci Rep, 6, 2016
|
|
4K0G
 
 | | Crystal structure of human CLIC1 C24S mutant | | Descriptor: | ACETATE ION, CALCIUM ION, Chloride intracellular channel protein 1 | | Authors: | Phang, J.M, Harrop, S.J, Duff, A.P, Wilk, K.E, Curmi, P.M.G. | | Deposit date: | 2013-04-03 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure analysis of CLIC1 C24 mutants
To be Published
|
|
4KGA
 
 | | Crystal structure of kallikrein-related peptidase 4 | | Descriptor: | 1,2-ETHANEDIOL, Kallikrein-4, NICKEL (II) ION | | Authors: | Ilyichova, O.V, Swedberg, J.E, de Veer, S.J, Sit, K.C, Harris, J.M, Buckle, A.M. | | Deposit date: | 2013-04-29 | | Release date: | 2014-04-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Direct and indirect mechanisms of KLK4 inhibition revealed by structure and dynamics
Sci Rep, 6, 2016
|
|
183D
 
 | | X-RAY STRUCTURE OF A DNA DECAMER CONTAINING 7, 8-DIHYDRO-8-OXOGUANINE | | Descriptor: | DNA (5'-D(*CP*CP*AP*(8OG)P*CP*GP*CP*TP*GP*G)-3') | | Authors: | Lipscomb, L.A, Peek, M.E, Morningstar, M.L, Verghis, S.M, Miller, E.M, Rich, A, Essigmann, J.M, Williams, L.D. | | Deposit date: | 1994-08-01 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray structure of a DNA decamer containing 7,8-dihydro-8-oxoguanine.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
3SS8
 
 | | Crystal structure of NFeoB from S. thermophilus bound to GDP.AlF4- and K+ | | Descriptor: | Ferrous iron uptake transporter protein B, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ash, M.R, Maher, M.J, Guss, J.M, Jormakka, M. | | Deposit date: | 2011-07-08 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | The initiation of GTP hydrolysis by the G-domain of FeoB: insights from a transition-state complex structure
Plos One, 6, 2011
|
|
1B79
 
 | |
4K8J
 
 | | Crystal Structure of Staphylococcal nuclease mutant V23L/V66I | | Descriptor: | Thermonuclease | | Authors: | Sanders, J.M, Gill, E, Roeser, J.R, Janowska, K, Sakon, J, Stites, W.E. | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hydrophobic core mutants of Staphylococcal nuclease
To be Published
|
|
