1DQ5
 
 | | Manganese;Manganese concanavalin A at pH 5.0 | | Descriptor: | Concanavalin-Br, MANGANESE (II) ION | | Authors: | Bouckaert, J, Dewallef, Y, Poortmans, F, Wyns, L, Loris, R. | | Deposit date: | 1999-12-30 | | Release date: | 2000-01-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural features of concanavalin A governing non-proline peptide isomerization
J.Biol.Chem., 275, 2000
|
|
3WRR
 
 | |
6I9J
 
 | | Human transforming growth factor beta2 in a tetragonal crystal form | | Descriptor: | Transforming growth factor beta-2 proprotein | | Authors: | Gomis-Ruth, F.X, Marino-Puertas, L, del Amo-Maestro, L, Goulas, T. | | Deposit date: | 2018-11-23 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recombinant production, purification, crystallization, and structure analysis of human transforming growth factor beta 2 in a new conformation.
Sci Rep, 9, 2019
|
|
1DBS
 
 | |
7CHS
 
 | | Crystal structure of SARS-CoV-2 antibody P22A-1D1 with RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P22A-1D1 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Ge, J, Wang, R, Zhang, Q. | | Deposit date: | 2020-07-06 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Potent and protective IGHV3-53/3-66 public antibodies and their shared escape mutant on the spike of SARS-CoV-2.
Nat Commun, 12, 2021
|
|
7CHO
 
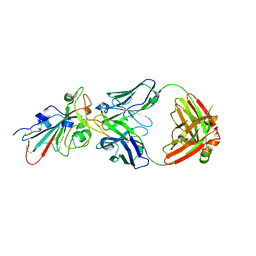 | | Crystal structure of SARS-CoV-2 antibody P5A-1D2 with RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P5A-1D2 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Ge, J, Wang, R, Zhang, Q. | | Deposit date: | 2020-07-06 | | Release date: | 2021-05-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.561 Å) | | Cite: | Potent and protective IGHV3-53/3-66 public antibodies and their shared escape mutant on the spike of SARS-CoV-2.
Nat Commun, 12, 2021
|
|
7CHP
 
 | | Crystal structure of SARS-CoV-2 antibody P5A-3C8 with RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P5A-3C8 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Ge, J, Wang, R, Zhang, Q. | | Deposit date: | 2020-07-06 | | Release date: | 2021-05-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.357 Å) | | Cite: | Potent and protective IGHV3-53/3-66 public antibodies and their shared escape mutant on the spike of SARS-CoV-2.
Nat Commun, 12, 2021
|
|
1UJ1
 
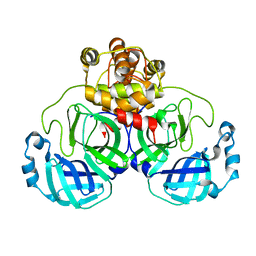 | | Crystal structure of SARS Coronavirus Main Proteinase (3CLpro) | | Descriptor: | 3C-like proteinase | | Authors: | Yang, H, Yang, M, Liu, Y, Bartlam, M, Ding, Y, Lou, Z, Sun, L, Zhou, Z, Ye, S, Anand, K, Pang, H, Gao, G.F, Hilgenfeld, R, Rao, Z. | | Deposit date: | 2003-07-25 | | Release date: | 2003-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1DFE
 
 | | NMR STRUCTURE OF RIBOSOMAL PROTEIN L36 FROM THERMUS THERMOPHILUS | | Descriptor: | L36 RIBOSOMAL PROTEIN, ZINC ION | | Authors: | Hard, T, Rak, A, Allard, P, Kloo, L, Garber, M. | | Deposit date: | 1999-11-19 | | Release date: | 1999-12-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of ribosomal protein L36 from Thermus thermophilus reveals a zinc-ribbon-like fold.
J.Mol.Biol., 296, 2000
|
|
1H97
 
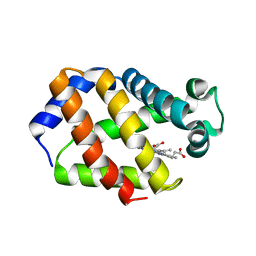 | | Trematode hemoglobin from Paramphistomum epiclitum | | Descriptor: | Globin-3, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Pesce, A, Dewilde, S, Kiger, L, Milani, M, Ascenzi, P, Marden, M.C, Van Hauwaert, M.L, Vanfleteren, J, Moens, L, Bolognesi, M. | | Deposit date: | 2001-03-02 | | Release date: | 2001-06-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Very High Resolution Structure of a Trematode Hemoglobin Displaying a Tyrb10-Tyre7 Heme Distal Residue Pair and High Oxygen Affinity
J.Mol.Biol., 309, 2001
|
|
1UK2
 
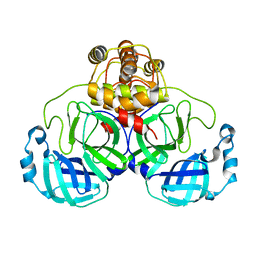 | | Crystal structure of SARS Coronavirus Main Proteinase (3CLpro) At pH8.0 | | Descriptor: | 3C-LIKE PROTEINASE | | Authors: | Yang, H, Yang, M, Liu, Y, Bartlam, M, Ding, Y, Lou, Z, Sun, L, Zhou, Z, Ye, S, Anand, K, Pang, H, Gao, G.F, Hilgenfeld, R, Rao, Z. | | Deposit date: | 2003-08-14 | | Release date: | 2003-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1UK3
 
 | | Crystal structure of SARS Coronavirus Main Proteinase (3CLpro) At pH7.6 | | Descriptor: | 3C-like proteinase | | Authors: | Yang, H, Yang, M, Liu, Y, Bartlam, M, Ding, Y, Lou, Z, Sun, L, Zhou, Z, Ye, S, Anand, K, Pang, H, Gao, G.F, Hilgenfeld, R, Rao, Z. | | Deposit date: | 2003-08-14 | | Release date: | 2003-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
6LOX
 
 | | Crystal Structure of human glutaminase with macrocyclic inhibitor | | Descriptor: | (E)-15,22-Dioxa-4,11-diaza-5(2,5)-thiadiazola-10(3,6)-pyridazina-1,14(1,3)-dibenzenacyclodocosaphan-18-ene-3,12-dione, Glutaminase kidney isoform, mitochondrial | | Authors: | Bian, J, Li, Z, Xu, X, Wang, J, Li, L. | | Deposit date: | 2020-01-07 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure-Enabled Discovery of Novel Macrocyclic Inhibitors Targeting Glutaminase 1 Allosteric Binding Site.
J.Med.Chem., 64, 2021
|
|
1UK4
 
 | | Crystal structure of SARS Coronavirus Main Proteinase (3CLpro) Complexed With An Inhibitor | | Descriptor: | 3C-like proteinase nsp5, 5-mer peptide of inhibitor | | Authors: | Yang, H, Yang, M, Liu, Y, Bartlam, M, Ding, Y, Lou, Z, Sun, L, Zhou, Z, Ye, S, Anand, K, Pang, H, Gao, G.F, Hilgenfeld, R, Rao, Z. | | Deposit date: | 2003-08-14 | | Release date: | 2003-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
2XTD
 
 | | Structure of the TBL1 tetramerisation domain | | Descriptor: | TBL1 F-BOX-LIKE/WD REPEAT-CONTAINING PROTEIN TBL1X | | Authors: | Oberoi, J, Fairall, L, Watson, P.J, Greenwood, J.A, Schwabe, J.W.R. | | Deposit date: | 2010-10-06 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis for the Assembly of the Smrt/Ncor Core Transcriptional Repression Machinery.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2H8F
 
 | | Crystal structure of deoxy hemoglobin from Trematomus bernacchii at pH 6.2 | | Descriptor: | Hemoglobin alpha subunit, Hemoglobin beta subunit, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mazzarella, L, Vergara, A, Vitagliano, L, Merlino, A, Bonomi, G, Scala, S, Verde, C, di Prisco, G. | | Deposit date: | 2006-06-07 | | Release date: | 2006-08-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High resolution crystal structure of deoxy hemoglobin from Trematomus bernacchii at different pH values: The role of histidine residues in modulating the strength of the root effect.
Proteins, 65, 2006
|
|
1DK7
 
 | |
1DK4
 
 | | CRYSTAL STRUCTURE OF MJ0109 GENE PRODUCT INOSITOL MONOPHOSPHATASE | | Descriptor: | INOSITOL MONOPHOSPHATASE, PHOSPHATE ION, ZINC ION | | Authors: | Stec, B, Yang, H, Johnson, K.A, Chen, L, Roberts, M.F. | | Deposit date: | 1999-12-06 | | Release date: | 2000-11-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | MJ0109 is an enzyme that is both an inositol monophosphatase and the 'missing' archaeal fructose-1,6-bisphosphatase.
Nat.Struct.Biol., 7, 2000
|
|
1DA2
 
 | | MOLECULAR AND CRYSTAL STRUCTURE OF D(CGCGMO4CG): N4-METHOXYCYTOSINE/GUANINE BASE-PAIRS IN Z-DNA | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*(C45)P*G)-3') | | Authors: | Van Meervelt, L, Moore, M.H, Lin, P.K.T, Brown, D.M, Kennard, O. | | Deposit date: | 1992-10-17 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular and crystal structure of d(CGCGmo4CG): N4-methoxycytosine.guanine base-pairs in Z-DNA.
J.Mol.Biol., 216, 1990
|
|
1DLY
 
 | | X-RAY CRYSTAL STRUCTURE OF HEMOGLOBIN FROM THE GREEN UNICELLULAR ALGA CHLAMYDOMONAS EUGAMETOS | | Descriptor: | 1,2-ETHANEDIOL, CYANIDE ION, HEMOGLOBIN, ... | | Authors: | Pesce, A, Couture, M, Guertin, M, Dewilde, S, Moens, L, Bolognesi, M. | | Deposit date: | 1999-12-13 | | Release date: | 2000-09-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel two-over-two alpha-helical sandwich fold is characteristic of the truncated hemoglobin family.
EMBO J., 19, 2000
|
|
1DI8
 
 | | THE STRUCTURE OF CYCLIN-DEPENDENT KINASE 2 (CDK2) IN COMPLEX WITH 4-[3-HYDROXYANILINO]-6,7-DIMETHOXYQUINAZOLINE | | Descriptor: | 4-[3-HYDROXYANILINO]-6,7-DIMETHOXYQUINAZOLINE, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Shewchuk, L, Hassell, A, Kuyper, L.F. | | Deposit date: | 1999-11-29 | | Release date: | 2000-11-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Binding mode of the 4-anilinoquinazoline class of protein kinase inhibitor: X-ray crystallographic studies of 4-anilinoquinazolines bound to cyclin-dependent kinase 2 and p38 kinase.
J.Med.Chem., 43, 2000
|
|
1DJ0
 
 | | THE CRYSTAL STRUCTURE OF E. COLI PSEUDOURIDINE SYNTHASE I AT 1.5 ANGSTROM RESOLUTION | | Descriptor: | CHLORIDE ION, PSEUDOURIDINE SYNTHASE I | | Authors: | Foster, P.G, Huang, L, Santi, D.V, Stroud, R.M. | | Deposit date: | 1999-11-30 | | Release date: | 2000-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structural basis for tRNA recognition and pseudouridine formation by pseudouridine synthase I.
Nat.Struct.Biol., 7, 2000
|
|
1DJP
 
 | | CRYSTAL STRUCTURE OF PSEUDOMONAS 7A GLUTAMINASE-ASPARAGINASE WITH THE INHIBITOR DON COVALENTLY BOUND IN THE ACTIVE SITE | | Descriptor: | 5,5-dihydroxy-6-oxo-L-norleucine, GLUTAMINASE-ASPARAGINASE | | Authors: | Ortlund, E, Lacount, M.W, Lewinski, K, Lebioda, L. | | Deposit date: | 1999-12-03 | | Release date: | 2000-01-24 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Reactions of Pseudomonas 7A glutaminase-asparaginase with diazo analogues of glutamine and asparagine result in unexpected covalent inhibitions and suggests an unusual catalytic triad Thr-Tyr-Glu.
Biochemistry, 39, 2000
|
|
2YHO
 
 | | The IDOL-UBE2D complex mediates sterol-dependent degradation of the LDL receptor | | Descriptor: | ACETATE ION, E3 UBIQUITIN-PROTEIN LIGASE MYLIP, UBIQUITIN-CONJUGATING ENZYME E2 D1, ... | | Authors: | Fairall, L, Goult, B.T, Millard, C.J, Tontonoz, P, Schwabe, J.W.R. | | Deposit date: | 2011-05-04 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Idol-Ube2D Complex Mediates Sterol-Dependent Degradation of the Ldl Receptor.
Genes Dev., 25, 2011
|
|
1DC9
 
 | | PROPERTIES AND CRYSTAL STRUCTURE OF A BETA-BARREL FOLDING MUTANT, V60N INTESTINAL FATTY ACID BINDING PROTEIN (IFABP) | | Descriptor: | INTESTINAL FATTY ACID BINDING PROTEIN | | Authors: | Ropson, I.J, Yowler, B.C, Dalessio, P.M, Banaszak, L, Thompson, J. | | Deposit date: | 1999-11-04 | | Release date: | 2000-03-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Properties and crystal structure of a beta-barrel folding mutant.
Biophys.J., 78, 2000
|
|
