8KD3
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification, H3K9Q mutation and 187bp DNA | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KD4
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification and 187bp DNA, class1 | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KD6
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification and 187bp DNA, class3 | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KC7
 
 | | Rpd3S histone deacetylase complex | | Descriptor: | Chromatin modification-related protein EAF3, Histone deacetylase RPD3, Transcriptional regulatory protein RCO1, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-06 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KD2
 
 | | Rpd3S in complex with 187bp nucleosome | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KD7
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification and 167bp DNA | | Descriptor: | 167bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Chang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
6WEL
 
 | | Structure of cGMP-unbound F403V/V407A mutant TAX-4 reconstituted in lipid nanodiscs | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cyclic nucleotide-gated cation channel, ... | | Authors: | Zheng, X, Fu, Z, Su, D, Zhang, Y, Li, M, Pan, Y, Li, H, Li, S, Grassucci, R.A, Ren, Z, Hu, Z, Li, X, Zhou, M, Li, G, Frank, J, Yang, J. | | Deposit date: | 2020-04-02 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Mechanism of ligand activation of a eukaryotic cyclic nucleotide-gated channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VYH
 
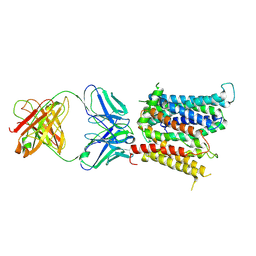 | | Cryo-EM structure of SLC40/ferroportin in complex with Fab | | Descriptor: | 11F9 heavy-chain, 11F9 light-chain, COBALT (II) ION, ... | | Authors: | Shen, J, Ren, Z, Pan, Y, Gao, S, Yan, N, Zhou, M. | | Deposit date: | 2020-02-26 | | Release date: | 2020-11-11 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of ion transport and inhibition in ferroportin.
Nat Commun, 11, 2020
|
|
6WEJ
 
 | | Structure of cGMP-unbound WT TAX-4 reconstituted in lipid nanodiscs | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cyclic nucleotide-gated cation channel, ... | | Authors: | Zheng, X, Fu, Z, Su, D, Zhang, Y, Li, M, Pan, Y, Li, H, Li, S, Grassucci, R.A, Ren, Z, Hu, Z, Li, X, Zhou, M, Li, G, Frank, J, Yang, J. | | Deposit date: | 2020-04-02 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Mechanism of ligand activation of a eukaryotic cyclic nucleotide-gated channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6WEK
 
 | | Structure of cGMP-bound WT TAX-4 reconstituted in lipid nanodiscs | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CYCLIC GUANOSINE MONOPHOSPHATE, ... | | Authors: | Zheng, X, Fu, Z, Su, D, Zhang, Y, Li, M, Pan, Y, Li, H, Li, S, Grassucci, R.A, Ren, Z, Hu, Z, Li, X, Zhou, M, Li, G, Frank, J, Yang, J. | | Deposit date: | 2020-04-02 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mechanism of ligand activation of a eukaryotic cyclic nucleotide-gated channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5HX7
 
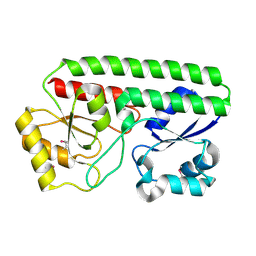 | | Metal ABC transporter from Listeria monocytogenes | | Descriptor: | Manganese-binding lipoprotein MntA | | Authors: | Osipiuk, J, Zhou, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-29 | | Release date: | 2016-02-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Metal ABC transporter from Listeria monocytogenes
to be published
|
|
5I4K
 
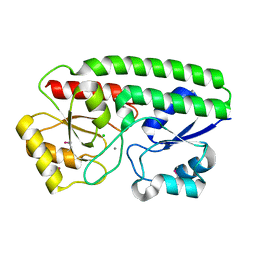 | | Metal ABC transporter from Listeria monocytogenes with manganese | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, Manganese-binding lipoprotein MntA | | Authors: | Osipiuk, J, Zhou, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-02-12 | | Release date: | 2016-02-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Metal ABC transporter from Listeria monocytogenes with manganese
to be published
|
|
8DL8
 
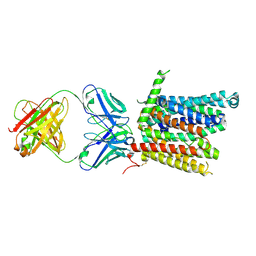 | | Cryo-EM structure of human ferroportin/slc40 bound to Co2+ in nanodisc | | Descriptor: | 11F9 heavy-chain, 11F9 light-chain, COBALT (II) ION, ... | | Authors: | Shen, J, Wilbon, A.S, Pan, Y, Zhou, M. | | Deposit date: | 2022-07-07 | | Release date: | 2022-12-07 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of ferroportin inhibition by minihepcidin PR73.
Plos Biol., 21, 2023
|
|
8DL7
 
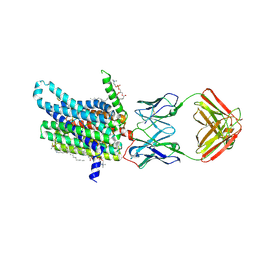 | | Cryo-EM structure of human ferroportin/slc40 bound to minihepcidin PR73 in nanodisc | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 11F9 heavy-chain, 11F9 light-chain, ... | | Authors: | Shen, J, Wilbon, A.S, Pan, Y, Zhou, M. | | Deposit date: | 2022-07-07 | | Release date: | 2022-12-07 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of ferroportin inhibition by minihepcidin PR73.
Plos Biol., 21, 2023
|
|
8DL6
 
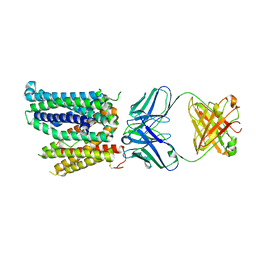 | | Cryo-EM structure of human ferroportin/slc40 bound to Ca2+ in nanodisc | | Descriptor: | 11F9 heavy-chain, 11F9 light-chain, CALCIUM ION, ... | | Authors: | Shen, J, Wilbon, A.S, Pan, Y, Zhou, M. | | Deposit date: | 2022-07-07 | | Release date: | 2023-01-25 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanism of Ca 2+ transport by ferroportin.
Elife, 12, 2023
|
|
7TH8
 
 | | Chickpea (Cicer arientinum) nodule-specific cysteine-rich peptide NCR13: Solution NMR structure of the isomer with C4:C10, C15:C30, and C23:C28 disulfide bonds | | Descriptor: | Nodule cysteine-rich protein 13 | | Authors: | Buchko, G.W, Zhou, M, Shah, D.M, Velivelli, S.L.S. | | Deposit date: | 2022-01-10 | | Release date: | 2022-02-02 | | Last modified: | 2025-01-08 | | Method: | SOLUTION NMR | | Cite: | Chickpea NCR13 disulfide cross-linking variants exhibit profound differences in antifungal activity and modes of action.
Plos Pathog., 20, 2024
|
|
8ERP
 
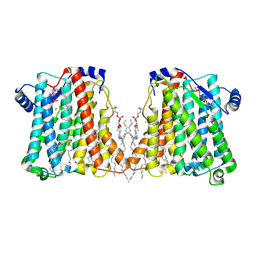 | | Structure of Xenopus cholinephosphotransferase1 in complex with CDP-choline | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Cholinephosphotransferase 1, MAGNESIUM ION, ... | | Authors: | Wang, L, Zhou, M. | | Deposit date: | 2022-10-12 | | Release date: | 2023-04-26 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of a eukaryotic cholinephosphotransferase-1 reveals mechanisms of substrate recognition and catalysis.
Nat Commun, 14, 2023
|
|
8ERO
 
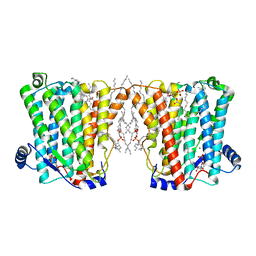 | | Structure of Xenopus cholinephosphotransferase1 in complex with CDP | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CYTIDINE-5'-DIPHOSPHATE, Cholinephosphotransferase 1, ... | | Authors: | Wang, L, Zhou, M. | | Deposit date: | 2022-10-12 | | Release date: | 2023-04-26 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of a eukaryotic cholinephosphotransferase-1 reveals mechanisms of substrate recognition and catalysis.
Nat Commun, 14, 2023
|
|
4YMK
 
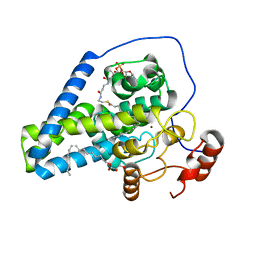 | | Crystal Structure of Stearoyl-Coenzyme A Desaturase 1 | | Descriptor: | Acyl-CoA desaturase 1, STEAROYL-COENZYME A, ZINC ION, ... | | Authors: | Bai, Y, McCoy, J.G, Rajashankar, K.R, Zhou, M. | | Deposit date: | 2015-03-06 | | Release date: | 2015-06-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | X-ray structure of a mammalian stearoyl-CoA desaturase.
Nature, 524, 2015
|
|
5IWS
 
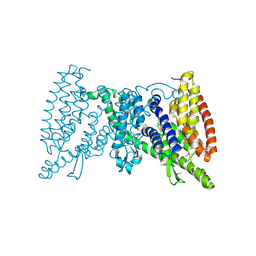 | | Crystal structure of the transporter MalT, the EIIC domain from the maltose-specific phosphotransferase system | | Descriptor: | Protein-N(Pi)-phosphohistidine-sugar phosphotransferase (Enzyme II of the phosphotransferase system) (PTS system glucose-specific IIBC component), alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | McCoy, J.G, Ren, Z, Levin, E.J, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2016-03-22 | | Release date: | 2016-05-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | The Structure of a Sugar Transporter of the Glucose EIIC Superfamily Provides Insight into the Elevator Mechanism of Membrane Transport.
Structure, 24, 2016
|
|
6VP0
 
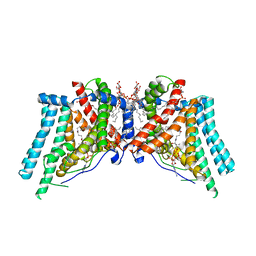 | | Human Diacylglycerol Acyltransferase 1 in complex with oleoyl-CoA | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Diacylglycerol O-acyltransferase 1, Lauryl Maltose Neopentyl Glycol, ... | | Authors: | Wang, L, Qian, H, Han, Y, Nian, Y, Ren, Z, Zhang, H, Hu, L, Prasad, B.V.V, Yan, N, Zhou, M. | | Deposit date: | 2020-02-01 | | Release date: | 2020-05-13 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure and mechanism of human diacylglycerol O-acyltransferase 1.
Nature, 581, 2020
|
|
6WF2
 
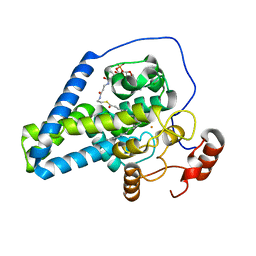 | | Crystal structure of mouse SCD1 with a diiron center | | Descriptor: | Acyl-CoA desaturase 1, FE (III) ION, S-{(3R,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14-dioxo-2,4,6-trioxa-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaheptadecan-17-yl} (9Z)-octadec-9-enethioate (non-preferred name) | | Authors: | Shen, J, Zhou, M. | | Deposit date: | 2020-04-03 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Structure and Mechanism of a Unique Diiron Center in Mammalian Stearoyl-CoA Desaturase.
J.Mol.Biol., 432, 2020
|
|
8UUK
 
 | | Pendrin in apo | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CHOLESTEROL, Lauryl Maltose Neopentyl Glycol, ... | | Authors: | Wang, L, Hoang, A, Zhou, M. | | Deposit date: | 2023-11-01 | | Release date: | 2024-02-07 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Mechanism of anion exchange and small-molecule inhibition of pendrin.
Nat Commun, 15, 2024
|
|
4R9X
 
 | | Crystal Structure of Putative Copper Homeostasis Protein CutC from Bacillus anthracis | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Copper homeostasis protein CutC, ... | | Authors: | Kim, Y, Zhou, M, Makowska-Grzyska, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-09-08 | | Release date: | 2014-09-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8515 Å) | | Cite: | Crystal Structure of Putative Copper Homeostasis Protein CutC
from Bacillus anthracis
To be Published, 2014
|
|
7TBS
 
 | | Crystal Structure of the Glutaredoxin 2 from Francisella tularensis | | Descriptor: | CHLORIDE ION, Glutaredoxin 2, SULFATE ION | | Authors: | Kim, Y, Zhou, M, Grimshaw, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-12-22 | | Release date: | 2022-01-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structure of the Glutaredoxin 2 from Francisella tularensis
To Be Published
|
|
