6J2H
 
 | | Crystal structure of bat (Pteropus Alecto) MHC class I Ptal-N*01:01 mutant (Met52 Asp53 Leu54 deleted) in complex with Hendra virus-derived peptide HeV1 | | Descriptor: | Beta-2-microglobulin, HeV1, Ptal-N*01:01 (Met52 Asp53 Leu54 deleted) | | Authors: | Lu, D, Liu, K.F, Yue, C, Lu, Q, Cheng, H, Chai, Y, Qi, J.X, Gao, G.F, Liu, W.J. | | Deposit date: | 2019-01-01 | | Release date: | 2019-09-18 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Peptide presentation by bat MHC class I provides new insight into the antiviral immunity of bats.
Plos Biol., 17, 2019
|
|
6IX2
 
 | | Structure of the A214C/A250I mutant of an epoxide hydrolase from Aspergillus usamii E001 (AuEH2) at 1.48 Angstroms resolution | | Descriptor: | GLYCEROL, Microsomal epoxide hyddrolase | | Authors: | Hu, D, Hu, B.C, Hou, X.D, Rao, Y.J, Wu, M.C. | | Deposit date: | 2018-12-09 | | Release date: | 2019-12-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.478 Å) | | Cite: | Structure-Guided Regulation in the Enantioselectivity of an Epoxide Hydrolase to Produce Enantiomeric Monosubstituted Epoxides and Vicinal Diols via Kinetic Resolution.
Org.Lett., 24, 2022
|
|
6J2F
 
 | | Crystal structure of bat (Pteropus Alecto) MHC class I Ptal-N*01:01 in complex with Hendra virus-derived peptide HeV2 | | Descriptor: | HeV2, Ptal-N*01:01, beta-2 microglobulin | | Authors: | Lu, D, Liu, K.F, Yue, C, Lu, Q, Cheng, H, Chai, Y, Qi, J.X, Gao, G.F, Liu, W.J. | | Deposit date: | 2019-01-01 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Peptide presentation by bat MHC class I provides new insight into the antiviral immunity of bats.
Plos Biol., 17, 2019
|
|
6J2I
 
 | | Crystal structure of bat (Pteropus Alecto) MHC class I Ptal-N*01:01 in complex with H17N10 influenza-like virus-derivrd peptide H17N10-NP | | Descriptor: | Beta-2-microglobulin, H17N10-NP, Ptal-N*01:01 | | Authors: | Lu, D, Liu, K.F, Yue, C, Lu, Q, Cheng, H, Chai, Y, Qi, J.X, Gao, G.F, Liu, W.J. | | Deposit date: | 2019-01-01 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Peptide presentation by bat MHC class I provides new insight into the antiviral immunity of bats.
Plos Biol., 17, 2019
|
|
6J2J
 
 | | Crystal structure of bat (Pteropus Alecto) MHC class I Ptal-N*01:01 in complex with MERS-CoV-derived peptide MERS-CoV-S3 | | Descriptor: | Beta-2-microglobulin, MERS-CoV-S3, Ptal-N*01:01 | | Authors: | Lu, D, Liu, K.F, Yue, C, Lu, Q, Cheng, H, Chai, Y, Qi, J.X, Gao, G.F, Liu, W.J. | | Deposit date: | 2019-01-01 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Peptide presentation by bat MHC class I provides new insight into the antiviral immunity of bats.
Plos Biol., 17, 2019
|
|
5GSN
 
 | | Tmm in complex with methimazole | | Descriptor: | 1-METHYL-1,3-DIHYDRO-2H-IMIDAZOLE-2-THIONE, FLAVIN-ADENINE DINUCLEOTIDE, Flavin-containing monooxygenase, ... | | Authors: | Zhang, Y.Z, Li, C.Y. | | Deposit date: | 2016-08-16 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Structural mechanism for bacterial oxidation of oceanic trimethylamine into trimethylamine N-oxide
Mol. Microbiol., 103, 2017
|
|
5TBP
 
 | | Crystal Structure of RXR-alpha ligand binding domain complexed with synthetic modulator K8003 | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Aleshin, A.E, Liddington, R.C, Su, Y, Zhang, X. | | Deposit date: | 2016-09-12 | | Release date: | 2017-08-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Modulation of nongenomic activation of PI3K signalling by tetramerization of N-terminally-cleaved RXR alpha.
Nat Commun, 8, 2017
|
|
2YIT
 
 | | Structural analysis of checkpoint kinase 2 in complex with PV1162, a novel inhibitor | | Descriptor: | N-{4-[(1E)-N-carbamimidoylbutanehydrazonoyl]phenyl}-5-methoxy-1H-indole-2-carboxamide, NITRATE ION, SERINE/THREONINE-PROTEIN KINASE CHK2 | | Authors: | Lountos, G.T, Jobson, A.G, Tropea, J.E, Self, C, Zhang, G, Pommier, Y, Shoemaker, R.H, Waugh, D.S. | | Deposit date: | 2011-05-16 | | Release date: | 2011-09-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-Ray Structures of Checkpoint Kinase 2 in Complex with Inhibitors that Target its Gatekeeper-Dependent Hydrophobic Pocket.
FEBS Lett., 585, 2011
|
|
2YIR
 
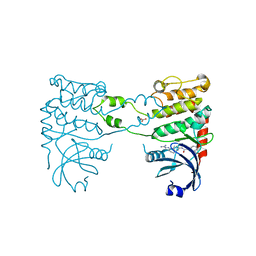 | | Structural analysis of checkpoint kinase 2 in complex with inhibitor PV1352 | | Descriptor: | (E)-N-(5-(2-CARBAMIMIDOYLHYDRAZONO)-5,6,7,8-TETRAHYDRONAPHTHALEN-2-YL)-7-NITRO-1H-INDOLE-2-CARBOXAMIDE, NITRATE ION, SERINE/THREONINE-PROTEIN KINASE CHK2 | | Authors: | Lountos, G.T, Jobson, A.G, Tropea, J.E, Self, C, Zhang, G, Pommier, Y, Shoemaker, R.H, Waugh, D.S. | | Deposit date: | 2011-05-16 | | Release date: | 2011-09-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-Ray Structures of Checkpoint Kinase 2 in Complex with Inhibitors that Target its Gatekeeper-Dependent Hydrophobic Pocket.
FEBS Lett., 585, 2011
|
|
5IPY
 
 | | Crystal structure of WT RnTmm | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-containing monooxygenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2016-03-10 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural mechanism for bacterial oxidation of oceanic trimethylamine into trimethylamine N-oxide
Mol. Microbiol., 103, 2017
|
|
5IQ4
 
 | | Crystal structure of RnTmm mutant Y207S soaking | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-containing monooxygenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, Y.Z, Li, C.Y. | | Deposit date: | 2016-03-10 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural mechanism for bacterial oxidation of oceanic trimethylamine into trimethylamine N-oxide
Mol. Microbiol., 103, 2017
|
|
5IQ1
 
 | | Crystal structure of RnTmm mutant Y207S | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-containing monooxygenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2016-03-10 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural mechanism for bacterial oxidation of oceanic trimethylamine into trimethylamine N-oxide
Mol. Microbiol., 103, 2017
|
|
5HX6
 
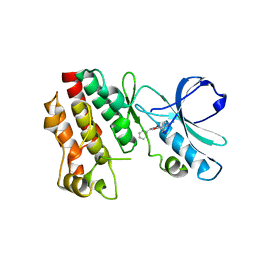 | | Crystal structure of RIP1 kinase with a benzo[b][1,4]oxazepin-4-one | | Descriptor: | 5-benzyl-N-[(3S)-5-methyl-4-oxo-2,3,4,5-tetrahydro-1,5-benzoxazepin-3-yl]-1,2-oxazole-3-carboxamide, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Campobasso, N, Ward, P. | | Deposit date: | 2016-01-29 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | DNA-Encoded Library Screening Identifies Benzo[b][1,4]oxazepin-4-ones as Highly Potent and Monoselective Receptor Interacting Protein 1 Kinase Inhibitors.
J.Med.Chem., 59, 2016
|
|
4JBA
 
 | |
4QHO
 
 | | Crystal structure of the human fat mass and obesity associated protein (FTO) in complex with CCO10 | | Descriptor: | Alpha-ketoglutarate-dependent dioxygenase FTO, N-{[3-hydroxy-6-(naphthalen-1-yl)pyridin-2-yl]carbonyl}glycine, ZINC ION | | Authors: | Aik, W.S, Clunie-O'Connor, C, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2014-05-28 | | Release date: | 2015-06-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure-Based Design of Selective Fat Mass and Obesity Associated Protein (FTO) Inhibitors.
J.Med.Chem., 64, 2021
|
|
6RLN
 
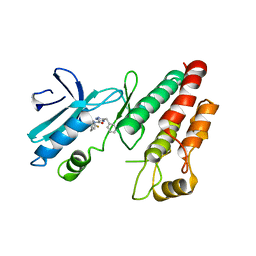 | | Crystal structure of RIP1 kinase in complex with GSK3145095 | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 1, ~{N}-[(3~{S})-7,9-bis(fluoranyl)-2-oxidanylidene-1,3,4,5-tetrahydro-1-benzazepin-3-yl]-3-(phenylmethyl)-1~{H}-1,2,4-triazole-5-carboxamide | | Authors: | Thorpe, J.H, Harris, P.A. | | Deposit date: | 2019-05-02 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Identification of a RIP1 Kinase Inhibitor Clinical Candidate (GSK3145095) for the Treatment of Pancreatic Cancer.
Acs Med.Chem.Lett., 10, 2019
|
|
2XK9
 
 | | Structural analysis of checkpoint kinase 2 (Chk2) in complex with inhibitor PV1533 | | Descriptor: | CHECKPOINT KINASE 2, N-{4-[(1E)-N-(N-hydroxycarbamimidoyl)ethanehydrazonoyl]phenyl}-7-nitro-1H-indole-2-carboxamide | | Authors: | Lountos, G.T, Jobson, A.G, Tropea, J.E, Self, C, Shoemaker, R.H, Pommier, Y, Waugh, D.S. | | Deposit date: | 2010-07-07 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Characterization of Inhibitor Complexes with Checkpoint Kinase 2 (Chk2), a Drug Target for Cancer Therapy.
J.Struct.Biol., 176, 2011
|
|
4G3Y
 
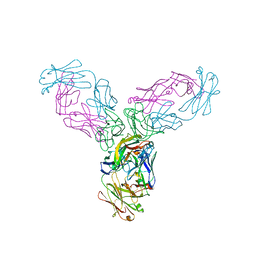 | | Crystal structure of TNF-alpha in complex with Infliximab Fab fragment | | Descriptor: | Tumor necrosis factor, infliximab Fab H, infliximab Fab L | | Authors: | Liang, S.Y, Dai, J.X, Guo, Y.J, Lou, Z.Y. | | Deposit date: | 2012-07-15 | | Release date: | 2013-03-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for treating tumor necrosis factor alpha (TNFalpha)-associated diseases with the therapeutic antibody infliximab
J.Biol.Chem., 288, 2013
|
|
3EOB
 
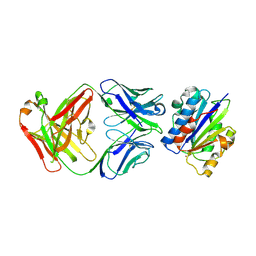 | |
3EOA
 
 | |
3EO9
 
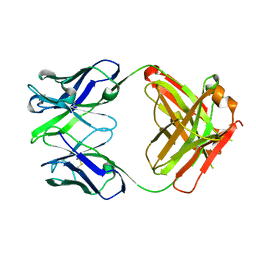 | | Crystal structure the Fab fragment of Efalizumab | | Descriptor: | Efalizumab Fab fragment, heavy chain, light chain | | Authors: | Li, S, Ding, J. | | Deposit date: | 2008-09-26 | | Release date: | 2009-04-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Efalizumab binding to the LFA-1 alphaL I domain blocks ICAM-1 binding via steric hindrance.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
8XJK
 
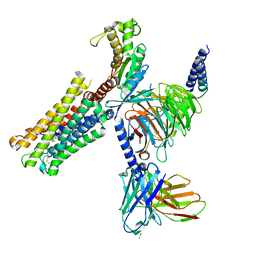 | | Cloprosetnol bound Prostaglandin F2-alpha receptor-Gq Protein Complex | | Descriptor: | (~{Z})-7-[(1~{R},2~{R},3~{R},5~{S})-2-[(~{E},3~{R})-4-(3-chloranylphenoxy)-3-oxidanyl-but-1-enyl]-3,5-bis(oxidanyl)cyclopentyl]hept-5-enoic acid, Antibody fragment scFv16, Engineered miniGq, ... | | Authors: | Zhang, X, Li, X, Liu, G, Gong, W. | | Deposit date: | 2023-12-21 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Structural basis for ligand recognition and activation of the prostanoid receptors.
Cell Rep, 43, 2024
|
|
8XJO
 
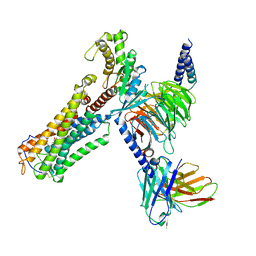 | | U46619 bound Thromboxane A2 receptor-Gq Protein Complex | | Descriptor: | (5Z)-7-{(1R,4S,5S,6R)-6-[(1E,3S)-3-hydroxyoct-1-en-1-yl]-2-oxabicyclo[2.2.1]hept-5-yl}hept-5-enoic acid, Antibody fragment scFv16, Engineered miniGq, ... | | Authors: | Zhang, X, Li, X, Liu, G, Gong, W. | | Deposit date: | 2023-12-21 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural basis for ligand recognition and activation of the prostanoid receptors.
Cell Rep, 43, 2024
|
|
8XJN
 
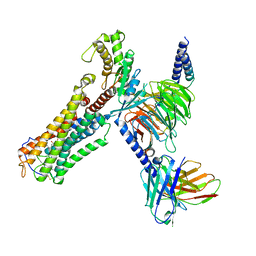 | | Cloprosetnol bound Thromboxane A2 receptor-Gq Protein Complex | | Descriptor: | (~{Z})-7-[(1~{R},2~{R},3~{R},5~{S})-2-[(~{E},3~{R})-4-(3-chloranylphenoxy)-3-oxidanyl-but-1-enyl]-3,5-bis(oxidanyl)cyclopentyl]hept-5-enoic acid, Antibody fragment scFv16, Engineered miniGq, ... | | Authors: | Zhang, X, Li, X, Liu, G, Gong, W. | | Deposit date: | 2023-12-21 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis for ligand recognition and activation of the prostanoid receptors.
Cell Rep, 43, 2024
|
|
8XJL
 
 | | PGF2-alpha bound Prostaglandin F2-alpha receptor-Gq Protein Complex | | Descriptor: | (Z)-7-[(1R,2R,3R,5S)-3,5-bis(oxidanyl)-2-[(E,3S)-3-oxidanyloct-1-enyl]cyclopentyl]hept-5-enoic acid, Antibody fragment scFv16, Engineered miniGq, ... | | Authors: | Zhang, X, Li, X, Liu, G, Gong, W. | | Deposit date: | 2023-12-21 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural basis for ligand recognition and activation of the prostanoid receptors.
Cell Rep, 43, 2024
|
|
