8J6M
 
 | | SIDT1 protein | | Descriptor: | CHOLESTEROL, Green fluorescent protein,SID1 transmembrane family member 1, OLEIC ACID, ... | | Authors: | Zhang, J.T, Jiang, D.H. | | Deposit date: | 2023-04-26 | | Release date: | 2024-05-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural insights into double-stranded RNA recognition and transport by SID-1.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7YB7
 
 | | anti-apoptotic protein BCL-2-M12 | | Descriptor: | Apoptosis regulator Bcl-2,Bcl-2-like protein 1, N-(2-acetamidoethyl)-4-(4,5-dihydro-1,3-thiazol-2-yl)benzamide, cp2 peptide | | Authors: | Li, F.W, Liu, C, Wu, D.L. | | Deposit date: | 2022-06-29 | | Release date: | 2023-11-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cyclic peptides discriminate BCL-2 and its clinical mutants from BCL-X L by engaging a single-residue discrepancy.
Nat Commun, 15, 2024
|
|
7U8O
 
 | | Structure of porcine V-ATPase with mEAK7 and SidK, Rotary state 2 | | Descriptor: | ATPase H(+)-transporting lysosomal accessory protein 2, ATPase H+ transporting accessory protein 1, Bacterial effector protein SidK, ... | | Authors: | Tan, Y.Z, Keon, K.A. | | Deposit date: | 2022-03-09 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | CryoEM of endogenous mammalian V-ATPase interacting with the TLDc protein mEAK-7.
Life Sci Alliance, 5, 2022
|
|
7U8R
 
 | | Structure of porcine kidney V-ATPase with SidK, Rotary State 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase H(+)-transporting lysosomal accessory protein 2, ATPase H+ transporting accessory protein 1, ... | | Authors: | Tan, Y.Z, Keon, K.A. | | Deposit date: | 2022-03-09 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | CryoEM of endogenous mammalian V-ATPase interacting with the TLDc protein mEAK-7.
Life Sci Alliance, 5, 2022
|
|
7U8Q
 
 | | Structure of porcine kidney V-ATPase with SidK, Rotary State 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase H(+)-transporting lysosomal accessory protein 2, ATPase H+ transporting accessory protein 1, ... | | Authors: | Tan, Y.Z, Keon, K.A. | | Deposit date: | 2022-03-09 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | CryoEM of endogenous mammalian V-ATPase interacting with the TLDc protein mEAK-7.
Life Sci Alliance, 5, 2022
|
|
7U8P
 
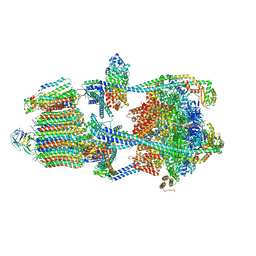 | | Structure of porcine kidney V-ATPase with SidK, Rotary State 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase H(+)-transporting lysosomal accessory protein 2, ATPase H+ transporting accessory protein 1, ... | | Authors: | Tan, Y.Z, Keon, K.A. | | Deposit date: | 2022-03-09 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | CryoEM of endogenous mammalian V-ATPase interacting with the TLDc protein mEAK-7.
Life Sci Alliance, 5, 2022
|
|
6M4W
 
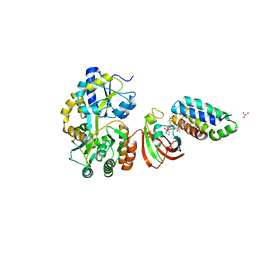 | | Crystal structure of MBP fused split FKBP-FRB T2098L mutant in complex with rapamycin | | Descriptor: | GLYCEROL, Peptidyl-prolyl cis-trans isomerase FKBP1A, RAPAMYCIN IMMUNOSUPPRESSANT DRUG, ... | | Authors: | Kikuchi, M, Wu, D, Inoue, T, Umehara, T. | | Deposit date: | 2020-03-09 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Rational design and implementation of a chemically inducible heterotrimerization system.
Nat.Methods, 17, 2020
|
|
6SO5
 
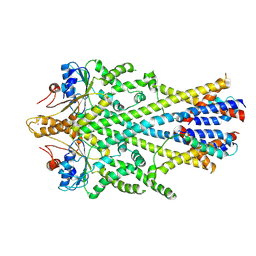 | | Homo sapiens WRB/CAML heterotetramer in complex with a TRC40 dimer | | Descriptor: | ATPase ASNA1, Calcium signal-modulating cyclophilin ligand, Tail-anchored protein insertion receptor WRB, ... | | Authors: | McDowell, M.A, Heimes, M, Wild, K, Flemming, D, Sinning, I. | | Deposit date: | 2019-08-29 | | Release date: | 2020-09-09 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural Basis of Tail-Anchored Membrane Protein Biogenesis by the GET Insertase Complex.
Mol.Cell, 80, 2020
|
|
7KDT
 
 | |
6RLN
 
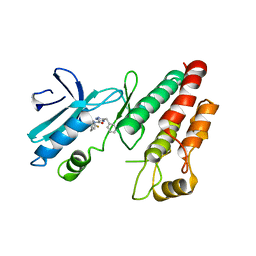 | | Crystal structure of RIP1 kinase in complex with GSK3145095 | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 1, ~{N}-[(3~{S})-7,9-bis(fluoranyl)-2-oxidanylidene-1,3,4,5-tetrahydro-1-benzazepin-3-yl]-3-(phenylmethyl)-1~{H}-1,2,4-triazole-5-carboxamide | | Authors: | Thorpe, J.H, Harris, P.A. | | Deposit date: | 2019-05-02 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Identification of a RIP1 Kinase Inhibitor Clinical Candidate (GSK3145095) for the Treatment of Pancreatic Cancer.
Acs Med.Chem.Lett., 10, 2019
|
|
8GRA
 
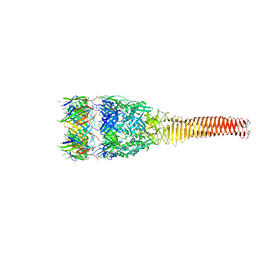 | |
6WM4
 
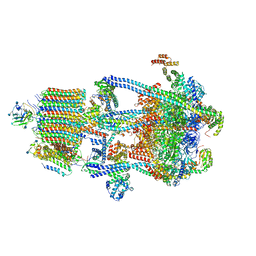 | | Human V-ATPase in state 3 with SidK and ADP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, Renin receptor, ... | | Authors: | Wang, L, Wu, H, Fu, T.M. | | Deposit date: | 2020-04-20 | | Release date: | 2020-11-11 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of a Complete Human V-ATPase Reveal Mechanisms of Its Assembly.
Mol.Cell, 80, 2020
|
|
6L8L
 
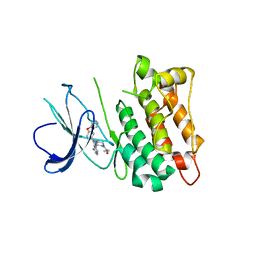 | | C-Src in complex with ibrutinib | | Descriptor: | 1-{(3R)-3-[4-amino-3-(4-phenoxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl}prop-2-en-1-one, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Guo, M, Dai, S, Chen, L, Chen, Y. | | Deposit date: | 2019-11-06 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.888 Å) | | Cite: | Characterization of ibrutinib as a non-covalent inhibitor of SRC-family kinases.
Bioorg.Med.Chem.Lett., 34, 2020
|
|
6M4J
 
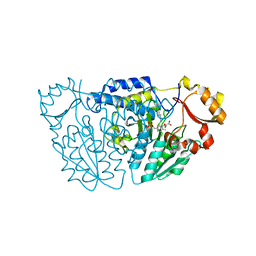 | | SspA in complex with cysteine | | Descriptor: | CYSTEINE, PYRIDOXAL-5'-PHOSPHATE, SspA complex protein | | Authors: | Liu, L, Gao, H. | | Deposit date: | 2020-03-07 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Analysis of an l-Cysteine Desulfurase from an Ssp DNA Phosphorothioation System.
Mbio, 11, 2020
|
|
7Y90
 
 | | Crystal Structure Analysis of cp1 bound BCL2 | | Descriptor: | (2R)-3-[2-(aminomethyl)-3-azanyl-1-[4-[2-(2-chloranylethanoylamino)ethylcarbamoyl]phenyl]prop-1-enyl]sulfanyl-2-(carboxyamino)propanoic acid, Apoptosis regulator Bcl-2, cp1 peptide | | Authors: | Li, F.W. | | Deposit date: | 2022-06-24 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Cyclic peptides discriminate BCL-2 and its clinical mutants from BCL-X L by engaging a single-residue discrepancy.
Nat Commun, 15, 2024
|
|
5ID6
 
 | | Structure of Cpf1/RNA Complex | | Descriptor: | Cpf1, MAGNESIUM ION, RNA (5'-R(P*AP*AP*UP*UP*UP*CP*UP*AP*CP*UP*AP*AP*GP*UP*GP*UP*AP*GP*AP*UP*C)-3') | | Authors: | Dong, D, Ren, K, Qiu, X, Wang, J, Huang, Z. | | Deposit date: | 2016-02-24 | | Release date: | 2016-04-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.382 Å) | | Cite: | The crystal structure of Cpf1 in complex with CRISPR RNA
Nature, 532, 2016
|
|
5Y8W
 
 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 1,2-ETHANEDIOL, 5-bromanyl-2-methoxy-N-(3-methyl-6-oxidanyl-1,2-benzoxazol-5-yl)benzenesulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Song, M, Wang, C. | | Deposit date: | 2017-08-21 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure-Based Discovery and Optimization of Benzo[ d]isoxazole Derivatives as Potent and Selective BET Inhibitors for Potential Treatment of Castration-Resistant Prostate Cancer (CRPC)
J. Med. Chem., 61, 2018
|
|
5Y94
 
 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 5-bromanyl-2-methoxy-N-[3-methyl-6-(methylamino)-1,2-benzoxazol-5-yl]benzenesulfonamide, Bromodomain-containing protein 4, GLYCEROL, ... | | Authors: | Xu, Y, Zhang, Y, Song, M, Wang, C. | | Deposit date: | 2017-08-22 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Discovery and Optimization of Benzo[ d]isoxazole Derivatives as Potent and Selective BET Inhibitors for Potential Treatment of Castration-Resistant Prostate Cancer (CRPC)
J. Med. Chem., 61, 2018
|
|
5Y8Y
 
 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 1,2-ETHANEDIOL, 5-bromanyl-2-methoxy-N-(6-methoxy-3-methyl-1,2-benzoxazol-5-yl)benzenesulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Song, M, Wang, C. | | Deposit date: | 2017-08-21 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure-Based Discovery and Optimization of Benzo[ d]isoxazole Derivatives as Potent and Selective BET Inhibitors for Potential Treatment of Castration-Resistant Prostate Cancer (CRPC)
J. Med. Chem., 61, 2018
|
|
5Y8Z
 
 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 1,2-ETHANEDIOL, 5-bromanyl-N-(3,6-dimethyl-1,2-benzoxazol-5-yl)-2-methoxy-benzenesulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Song, M, Wang, C. | | Deposit date: | 2017-08-22 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure-Based Discovery and Optimization of Benzo[ d]isoxazole Derivatives as Potent and Selective BET Inhibitors for Potential Treatment of Castration-Resistant Prostate Cancer (CRPC)
J. Med. Chem., 61, 2018
|
|
5Y8C
 
 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 1,2-ETHANEDIOL, 5-chloranyl-2-methoxy-N-(6-methoxy-3-methyl-1,2-benzoxazol-5-yl)benzenesulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Song, M, Wang, C. | | Deposit date: | 2017-08-21 | | Release date: | 2018-06-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structure-Based Discovery and Optimization of Benzo[ d]isoxazole Derivatives as Potent and Selective BET Inhibitors for Potential Treatment of Castration-Resistant Prostate Cancer (CRPC)
J. Med. Chem., 61, 2018
|
|
9JFL
 
 | |
7YW0
 
 | | Bacteroides fragilis Hcp5 | | Descriptor: | Bacterodales T6SS protein TssD (Hcp) | | Authors: | Wen, Y, He, W, Bai, Y. | | Deposit date: | 2022-08-20 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure and assembly of type VI secretion system cargo delivery vehicle.
Cell Rep, 42, 2023
|
|
8Q3V
 
 | | Cryo-EM structure of the methanogenic Na+ translocating N5-methyl-H4MPT:CoM methyltransferase complex | | Descriptor: | MAGNESIUM ION, SODIUM ION, Tetrahydromethanopterin S-methyltransferase subunit A 1, ... | | Authors: | Aziz, I, Vonck, J, Ermler, U. | | Deposit date: | 2023-08-04 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.08 Å) | | Cite: | Structural and mechanistic basis of the central energy-converting methyltransferase complex of methanogenesis.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8Q54
 
 | | N5-methyl-H4MPT:CoM methyltransferase -coenzyme M complex + CoM | | Descriptor: | 1-THIOETHANESULFONIC ACID, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Aziz, I, Vonck, J, Ermler, U. | | Deposit date: | 2023-08-08 | | Release date: | 2024-08-21 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Structural and mechanistic basis of the central energy-converting methyltransferase complex of methanogenesis.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
