5WYJ
 
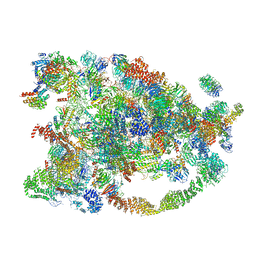 | | Cryo-EM structure of the 90S small subunit pre-ribosome (Dhr1-depleted, Enp1-TAP, state 1) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S ribosomal RNA, 40S ribosomal protein S1-A, ... | | Authors: | Ye, K, Zhu, X, Sun, Q. | | Deposit date: | 2017-01-13 | | Release date: | 2017-03-29 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | Molecular architecture of the 90S small subunit pre-ribosome.
Elife, 6, 2017
|
|
5WWM
 
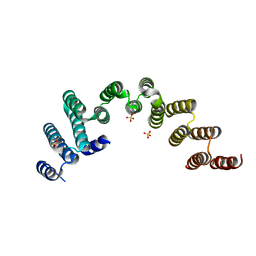 | | Crystal structure of the TPR domain of Rrp5 | | Descriptor: | SULFATE ION, rRNA biogenesis protein RRP5 | | Authors: | Ye, K, Chen, X. | | Deposit date: | 2017-01-03 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.809 Å) | | Cite: | Correction: Molecular architecture of the 90S small subunit pre-ribosome
Elife, 6, 2017
|
|
5WYL
 
 | |
5F9P
 
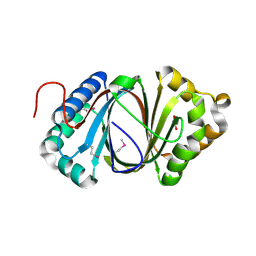 | | Crystal structure study of anthrone oxidase-like protein | | Descriptor: | Anthrone oxidase-like protein, GLYCEROL | | Authors: | Gao, X, Wu, D, Fan, K, Liu, Z.-J. | | Deposit date: | 2015-12-10 | | Release date: | 2016-12-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.078 Å) | | Cite: | Structure and Function of a C-C Bond Cleaving Oxygenase in Atypical Angucycline Biosynthesis
ACS Chem. Biol., 12, 2017
|
|
8YYP
 
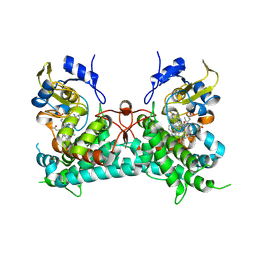 | | Crystal structure of PtmB in complex with cyclo-(L-Trp-L-Trp) and Adenine | | Descriptor: | (3S,6S)-3,6-bis[(1H-indol-3-yl)methyl]piperazine-2,5-dione, ADENINE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Zhang, Z.Y, Qu, X.D, Duan, B.R, Wei, G.Z. | | Deposit date: | 2024-04-04 | | Release date: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A nucleobase-driven P450 peroxidase system enables regio- and stereo-specific formation of C─C and C─N bonds.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7QNY
 
 | | The receptor binding domain of SARS-CoV-2 spike glycoprotein in complex with COVOX-58 and COVOX-158 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-158 heavy chain, COVOX-158 light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-12-23 | | Release date: | 2022-01-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses.
Cell, 185, 2022
|
|
7QNW
 
 | | The receptor binding domain of SARS-CoV-2 Omicron variant spike glycoprotein in complex with Beta-55 and EY6A Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Beta-55 heavy chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-12-23 | | Release date: | 2022-01-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses.
Cell, 185, 2022
|
|
7QNX
 
 | | The receptor binding domain of SARS-CoV-2 spike glycoprotein in complex with Beta-55 and EY6A Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-55 heavy chain, Beta-55 light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-12-23 | | Release date: | 2022-01-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses.
Cell, 185, 2022
|
|
7URF
 
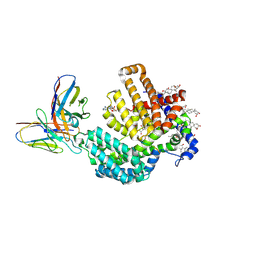 | | Human HHAT H379C in complex with SHH N-terminal peptide | | Descriptor: | 3H02 heavy chain, 3H02 light chain, Digitonin, ... | | Authors: | Liu, Y, Qi, X, Li, X. | | Deposit date: | 2022-04-21 | | Release date: | 2022-07-20 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanisms and inhibition of Porcupine-mediated Wnt acylation.
Nature, 607, 2022
|
|
7URE
 
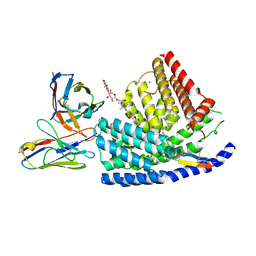 | | Human PORCN in complex with palmitoleoylated WNT3A peptide | | Descriptor: | 2C11 heavy chain, 2C11 light chain, Digitonin, ... | | Authors: | Liu, Y, Qi, X, Li, X. | | Deposit date: | 2022-04-21 | | Release date: | 2022-07-20 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Mechanisms and inhibition of Porcupine-mediated Wnt acylation.
Nature, 607, 2022
|
|
7URC
 
 | | Human PORCN in complex with LGK974 | | Descriptor: | 2-[(2P)-2',3-dimethyl[2,4'-bipyridin]-5-yl]-N-[(5P)-5-(pyrazin-2-yl)pyridin-2-yl]acetamide, 2C11 heavy chain, 2C11 light chain, ... | | Authors: | Liu, Y, Qi, X, Li, X. | | Deposit date: | 2022-04-21 | | Release date: | 2022-07-20 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Mechanisms and inhibition of Porcupine-mediated Wnt acylation.
Nature, 607, 2022
|
|
7URA
 
 | | Human PORCN in complex with Palmitoleoyl-CoA | | Descriptor: | 2C11 heavy chain, 2C11 light chain, CHOLESTEROL, ... | | Authors: | Liu, Y, Qi, X, Li, X. | | Deposit date: | 2022-04-21 | | Release date: | 2022-07-20 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Mechanisms and inhibition of Porcupine-mediated Wnt acylation.
Nature, 607, 2022
|
|
7URD
 
 | | Human PORCN in complex with LGK974 and WNT3A peptide | | Descriptor: | 2-[(2P)-2',3-dimethyl[2,4'-bipyridin]-5-yl]-N-[(5P)-5-(pyrazin-2-yl)pyridin-2-yl]acetamide, 2C11 heavy chain, 2C11 light chain, ... | | Authors: | Liu, Y, Qi, X, Li, X. | | Deposit date: | 2022-04-21 | | Release date: | 2022-07-20 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Mechanisms and inhibition of Porcupine-mediated Wnt acylation.
Nature, 607, 2022
|
|
4ZSE
 
 | |
7OYL
 
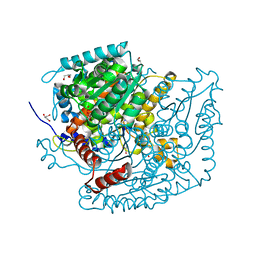 | | Phosphoglucose isomerase of Aspergillus fumigatus in complexed with Glucose-6-phosphate | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Raimi, O.G, Yan, K, Fang, W, van Aalten, D.M.F. | | Deposit date: | 2021-06-24 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Phosphoglucose Isomerase Is Important for Aspergillus fumigatus Cell Wall Biogenesis.
Mbio, 13, 2022
|
|
3SRQ
 
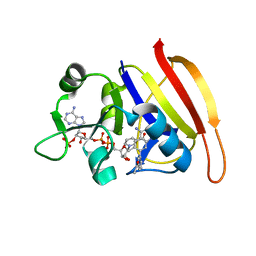 | | S. aureus Dihydrofolate Reductase complexed with novel 7-aryl-2,4-diaminoquinazolines | | Descriptor: | 1-[3-(2,4-diamino-6-methylquinazolin-7-yl)phenyl]ethanone, Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hilgers, M. | | Deposit date: | 2011-07-07 | | Release date: | 2011-08-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure-based design of new DHFR-based antibacterial agents: 7-aryl-2,4-diaminoquinazolines.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3SRS
 
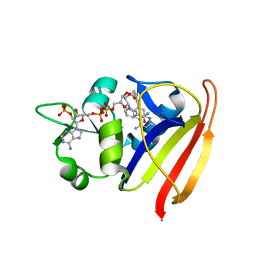 | | S. aureus Dihydrofolate Reductase complexed with novel 7-aryl-2,4-diaminoquinazolines | | Descriptor: | 7-(5-bromo-2-ethoxyphenyl)-6-methylquinazoline-2,4-diamine, Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hilgers, M. | | Deposit date: | 2011-07-07 | | Release date: | 2011-08-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based design of new DHFR-based antibacterial agents: 7-aryl-2,4-diaminoquinazolines.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
6LCY
 
 | | Crystal structure of Synaptotagmin-7 C2B in complex with IP6 | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Synaptotagmin-7 | | Authors: | Zhang, Y, Zhang, X, Rao, F, Wang, C. | | Deposit date: | 2019-11-20 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | 5-IP 7 is a GPCR messenger mediating neural control of synaptotagmin-dependent insulin exocytosis and glucose homeostasis.
Nat Metab, 3, 2021
|
|
8PTG
 
 | | Structure of the transcription termination factor Rho bound to RNA at the PBS and SBS | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Said, N, Hilal, T, Wahl, M.C. | | Deposit date: | 2023-07-14 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Sm-like protein Rof inhibits transcription termination factor rho by binding site obstruction and conformational insulation.
Nat Commun, 15, 2024
|
|
8PTM
 
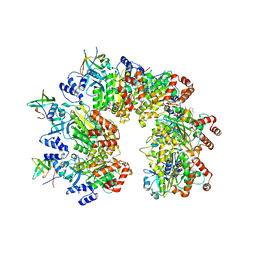 | | Structure of the transcription termination factor Rho in complex with Rof and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Protein rof, ... | | Authors: | Said, N, Hilal, T, Wahl, M.C. | | Deposit date: | 2023-07-14 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Sm-like protein Rof inhibits transcription termination factor rho by binding site obstruction and conformational insulation.
Nat Commun, 15, 2024
|
|
8PTP
 
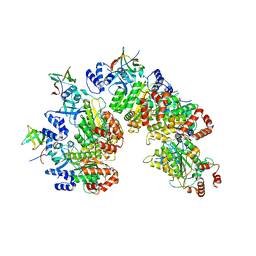 | | Structure of Rho pentamer in complex with Rof | | Descriptor: | Protein rof, Transcription termination factor Rho | | Authors: | Said, N, Hilal, T, Wahl, M.C. | | Deposit date: | 2023-07-14 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Sm-like protein Rof inhibits transcription termination factor rho by binding site obstruction and conformational insulation.
Nat Commun, 15, 2024
|
|
8PTN
 
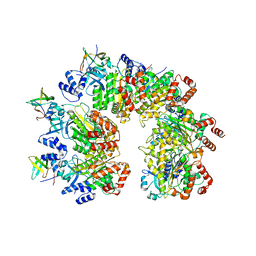 | |
8PTO
 
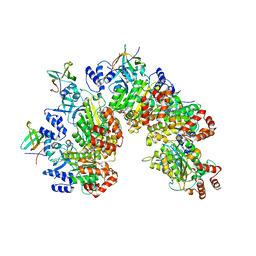 | | Structure of Rho pentamer in complex with Rof and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Protein rof, Transcription termination factor Rho | | Authors: | Said, N, Hilal, T, Wahl, M.C. | | Deposit date: | 2023-07-14 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Sm-like protein Rof inhibits transcription termination factor rho by binding site obstruction and conformational insulation.
Nat Commun, 15, 2024
|
|
3SRU
 
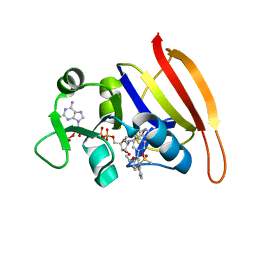 | | S. aureus Dihydrofolate Reductase complexed with novel 7-aryl-2,4-diaminoquinazolines | | Descriptor: | Dihydrofolate reductase, N-[3'-(2,4-diaminoquinazolin-7-yl)-4'-ethoxybiphenyl-3-yl]methanesulfonamide, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hilgers, M. | | Deposit date: | 2011-07-07 | | Release date: | 2011-08-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based design of new DHFR-based antibacterial agents: 7-aryl-2,4-diaminoquinazolines.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
6KJ9
 
 | | E. coli ATCase catalytic subunit mutant - G128/130A | | Descriptor: | Aspartate carbamoyltransferase catalytic subunit | | Authors: | Lei, Z, Zheng, J, Jia, Z.C. | | Deposit date: | 2019-07-22 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | New regulatory mechanism-based inhibitors of aspartate transcarbamoylase for potential anticancer drug development.
Febs J., 287, 2020
|
|
