1JL0
 
 | | Structure of a Human S-Adenosylmethionine Decarboxylase Self-processing Ester Intermediate and Mechanism of Putrescine Stimulation of Processing as Revealed by the H243A Mutant | | Descriptor: | 1,4-DIAMINOBUTANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, S-ADENOSYLMETHIONINE DECARBOXYLASE PROENZYME | | Authors: | Ekstrom, J.L, Tolbert, W.D, Xiong, H, Pegg, A.E, Ealick, S.E. | | Deposit date: | 2001-07-13 | | Release date: | 2001-08-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a human S-adenosylmethionine decarboxylase self-processing ester intermediate and mechanism of putrescine stimulation of processing as revealed by the H243A mutant.
Biochemistry, 40, 2001
|
|
3QPB
 
 | | Crystal Structure of Streptococcus Pyogenes Uridine Phosphorylase Reveals a Subclass of the NP-I Superfamily | | Descriptor: | 1-O-phosphono-alpha-D-ribofuranose, URACIL, Uridine phosphorylase | | Authors: | Tran, T.H, Christoffersen, S, Parker, W.B, Piskur, J, Serra, I, Terreni, M, Ealick, S.E. | | Deposit date: | 2011-02-11 | | Release date: | 2011-08-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The Crystal Structure of Streptococcus pyogenes Uridine Phosphorylase Reveals a Distinct Subfamily of Nucleoside Phosphorylases.
Biochemistry, 50, 2011
|
|
3QVK
 
 | | Allantoin racemase from Klebsiella pneumoniae | | Descriptor: | 1-(2,5-DIOXO-2,5-DIHYDRO-1H-IMIDAZOL-4-YL)UREA, Putative hydantoin racemase | | Authors: | French, J.B, Neau, D.B, Ealick, S.E. | | Deposit date: | 2011-02-25 | | Release date: | 2011-05-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Characterization of the Structure and Function of Klebsiella pneumoniae Allantoin Racemase.
J.Mol.Biol., 410, 2011
|
|
3QW4
 
 | | Structure of Leishmania donovani UMP synthase | | Descriptor: | CHLORIDE ION, UMP synthase, URIDINE-5'-MONOPHOSPHATE | | Authors: | French, J.B, Ealick, S.E. | | Deposit date: | 2011-02-26 | | Release date: | 2011-04-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Leishmania donovani UMP synthase is essential for promastigote viability and has an unusual tetrameric structure that exhibits substrate-controlled oligomerization.
J.Biol.Chem., 286, 2011
|
|
3QW3
 
 | | Structure of Leishmania donovani OMP decarboxylase | | Descriptor: | Orotidine-5-phosphate decarboxylase/orotate phosphoribosyltransferase, putative (Ompdcase-oprtase, putative), ... | | Authors: | French, J.B, Ealick, S.E. | | Deposit date: | 2011-02-26 | | Release date: | 2011-04-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Leishmania donovani UMP synthase is essential for promastigote viability and has an unusual tetrameric structure that exhibits substrate-controlled oligomerization.
J.Biol.Chem., 286, 2011
|
|
3RM5
 
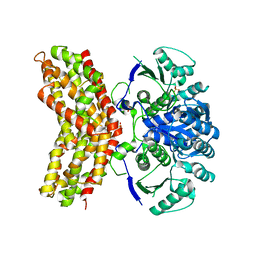 | |
3QVA
 
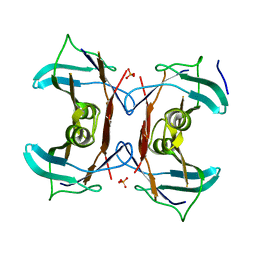 | |
3QVL
 
 | | Allantoin racemase from Klebsiella pneumoniae | | Descriptor: | Putative hydantoin racemase, [(4R)-2,5-dioxoimidazolidin-4-yl]acetic acid | | Authors: | French, J.B, Neau, D.B, Ealick, S.E. | | Deposit date: | 2011-02-25 | | Release date: | 2011-05-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.822 Å) | | Cite: | Characterization of the Structure and Function of Klebsiella pneumoniae Allantoin Racemase.
J.Mol.Biol., 410, 2011
|
|
3QVJ
 
 | |
3RW9
 
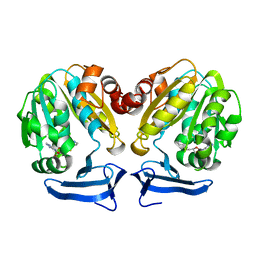 | | Crystal Structure of human Spermidine Synthase in Complex with decarboxylated S-adenosylhomocysteine | | Descriptor: | 5'-S-(3-aminopropyl)-5'-thioadenosine, Spermidine synthase | | Authors: | Seckute, J, McCloskey, D.E, Thomas, H.J, Secrist III, J.A, Pegg, A.E, Ealick, S.E. | | Deposit date: | 2011-05-08 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding and inhibition of human spermidine synthase by decarboxylated S-adenosylhomocysteine.
Protein Sci., 20, 2011
|
|
2NTM
 
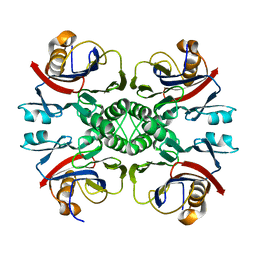 | |
3UAW
 
 | | Crystal structure of adenosine phosphorylase from Bacillus cereus complexed with adenosine | | Descriptor: | ADENOSINE, GLYCEROL, Purine nucleoside phosphorylase deoD-type, ... | | Authors: | Dessanti, P, Zhang, Y, Allegrini, S, Tozzi, M.G, Sgarrella, F, Ealick, S.E. | | Deposit date: | 2011-10-22 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural basis of the substrate specificity of Bacillus cereus adenosine phosphorylase.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2NTL
 
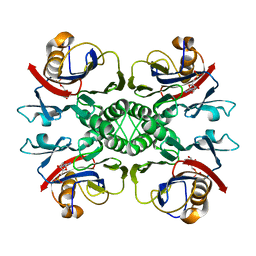 | | Crystal structure of PurO/AICAR from Methanothermobacter thermoautotrophicus | | Descriptor: | AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, IMP cyclohydrolase | | Authors: | Kang, Y.N, Tran, A, White, R.H, Ealick, S.E. | | Deposit date: | 2006-11-07 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A novel function for the N-terminal nucleophile hydrolase fold demonstrated by the structure of an archaeal inosine monophosphate cyclohydrolase.
Biochemistry, 46, 2007
|
|
2PMV
 
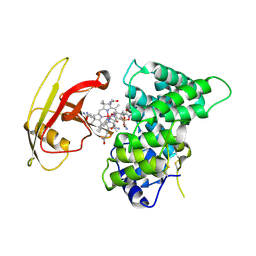 | | Crystal Structure of Human Intrinsic Factor- Cobalamin Complex at 2.6 A Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COBALAMIN, Gastric intrinsic factor | | Authors: | Mathews, F.S, Gordon, M.M, Chen, Z, Rajashankar, K.R, Ealick, S.E, Alpers, D.H, Sukumar, N. | | Deposit date: | 2007-04-23 | | Release date: | 2007-10-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of human intrinsic factor: Cobalamin complex at 2.6-A resolution
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
3RI6
 
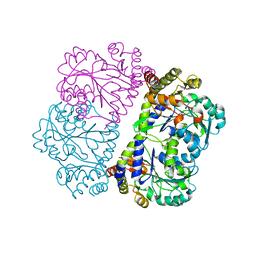 | | A Novel Mechanism of Sulfur Transfer Catalyzed by O-Acetylhomoserine Sulfhydrylase in Methionine Biosynthetic Pathway of Wolinella succinogenes | | Descriptor: | O-ACETYLHOMOSERINE SULFHYDRYLASE | | Authors: | Tran, T.H, Krishnamoorthy, K, Begley, T.P, Ealick, S.E. | | Deposit date: | 2011-04-13 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A novel mechanism of sulfur transfer catalyzed by O-acetylhomoserine sulfhydrylase in the methionine-biosynthetic pathway of Wolinella succinogenes.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3UAV
 
 | | Crystal structure of adenosine phosphorylase from Bacillus cereus | | Descriptor: | GLYCEROL, Purine nucleoside phosphorylase deoD-type, SULFATE ION | | Authors: | Dessanti, P, Zhang, Y, Allegrini, S, Tozzi, M.G, Sgarrella, F, Ealick, S.E. | | Deposit date: | 2011-10-22 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of the substrate specificity of Bacillus cereus adenosine phosphorylase.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3UAX
 
 | | Crystal structure of adenosine phosphorylase from Bacillus cereus complexed with inosine | | Descriptor: | GLYCEROL, INOSINE, Purine nucleoside phosphorylase deoD-type, ... | | Authors: | Dessanti, P, Zhang, Y, Allegrini, S, Tozzi, M.G, Sgarrella, F, Ealick, S.E. | | Deposit date: | 2011-10-22 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural basis of the substrate specificity of Bacillus cereus adenosine phosphorylase.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3UAY
 
 | | Crystal structure of Bacillus cereus adenosine phosphorylase D204N mutant complexed with adenosine | | Descriptor: | ADENOSINE, GLYCEROL, Purine nucleoside phosphorylase deoD-type, ... | | Authors: | Dessanti, P, Zhang, Y, Allegrini, S, Tozzi, M.G, Sgarrella, F, Ealick, S.E. | | Deposit date: | 2011-10-22 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of the substrate specificity of Bacillus cereus adenosine phosphorylase.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2NTK
 
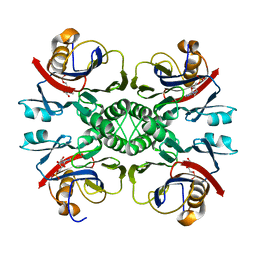 | | Crystal structure of PurO/IMP from Methanothermobacter thermoautotrophicus | | Descriptor: | IMP cyclohydrolase, INOSINIC ACID | | Authors: | Kang, Y.N, Tran, A, White, R.H, Ealick, S.E. | | Deposit date: | 2006-11-07 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A novel function for the N-terminal nucleophile hydrolase fold demonstrated by the structure of an archaeal inosine monophosphate cyclohydrolase.
Biochemistry, 46, 2007
|
|
2QCX
 
 | | Crystal structure of Bacillus subtilis TenA Y112F mutant complexed with formyl aminomethyl pyrimidine | | Descriptor: | N-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-N-(2-HYDROXYETHYL)FORMAMIDE, Transcriptional activator tenA | | Authors: | Zhang, Y, Jenkins, A.L, Begley, T.P, Ealick, S.E. | | Deposit date: | 2007-06-19 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutagenesis studies on TenA: A thiamin salvage enzyme from Bacillus subtilis
Bioorg.Chem., 36, 2008
|
|
3UAZ
 
 | | Crystal structure of Bacillus cereus adenosine phosphorylase D204N mutant complexed with inosine | | Descriptor: | GLYCEROL, INOSINE, Purine nucleoside phosphorylase deoD-type, ... | | Authors: | Dessanti, P, Zhang, Y, Allegrini, S, Tozzi, M.G, Sgarrella, F, Ealick, S.E. | | Deposit date: | 2011-10-22 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of the substrate specificity of Bacillus cereus adenosine phosphorylase.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2NOX
 
 | | Crystal structure of tryptophan 2,3-dioxygenase from Ralstonia metallidurans | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Tryptophan 2,3-dioxygenase | | Authors: | Zhang, Y, Kang, S.A, Mukherjee, T, Bale, S, Crane, B.R, Begley, T.P, Ealick, S.E. | | Deposit date: | 2006-10-26 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure and mechanism of tryptophan 2,3-dioxygenase, a heme enzyme involved in tryptophan catabolism and in quinolinate biosynthesis.
Biochemistry, 46, 2007
|
|
2R85
 
 | | Crystal structure of PurP from Pyrococcus furiosus complexed with AMP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Zhang, Y, White, R.H, Ealick, S.E. | | Deposit date: | 2007-09-10 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure and function of 5-formaminoimidazole-4-carboxamide ribonucleotide synthetase from Methanocaldococcus jannaschii.
Biochemistry, 47, 2008
|
|
3EP6
 
 | | Human AdoMetDC D174N mutant complexed with S-Adenosylmethionine methyl ester and no putrescine bound | | Descriptor: | PYRUVIC ACID, S-ADENOSYLMETHIONINE METHYL ESTER, S-adenosylmethionine decarboxylase alpha chain, ... | | Authors: | Bale, S, Lopez, M.M, Makhatadze, G.I, Fang, Q, Pegg, A.E, Ealick, S.E. | | Deposit date: | 2008-09-29 | | Release date: | 2008-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Putrescine Activation of Human S-Adenosylmethionine Decarboxylase.
Biochemistry, 47, 2008
|
|
3EP7
 
 | | Human AdoMetDC E256Q mutant complexed with S-Adenosylmethionine methyl ester and no putrescine bound | | Descriptor: | PYRUVIC ACID, S-ADENOSYLMETHIONINE METHYL ESTER, S-adenosylmethionine decarboxylase alpha chain, ... | | Authors: | Bale, S, Lopez, M.M, Makhatadze, G.I, Fang, Q, Pegg, A.E, Ealick, S.E. | | Deposit date: | 2008-09-29 | | Release date: | 2008-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Putrescine Activation of Human S-Adenosylmethionine Decarboxylase.
Biochemistry, 47, 2008
|
|
