4IHQ
 
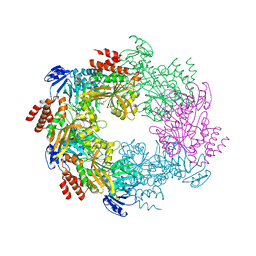 | | Archaellum Assembly ATPase FlaI bound to ADP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, FlaI ATPase, ... | | Authors: | Reindl, S, Williams, G.J, Tainer, J.A. | | Deposit date: | 2012-12-19 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into FlaI Functions in Archaeal Motor Assembly and Motility from Structures, Conformations, and Genetics.
Mol.Cell, 49, 2013
|
|
4IP4
 
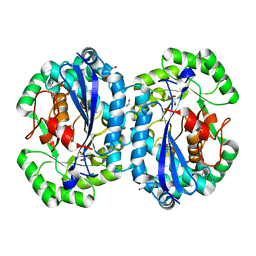 | | Crystal structure of l-fuconate dehydratase from Silicibacter sp. tm1040 liganded with Mg | | Descriptor: | CARBON DIOXIDE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Lukk, T, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-01-09 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.128 Å) | | Cite: | Crystal structure of l-fuconate dehydratase from Silicibacter sp. tm1040 liganded with mg
To be Published
|
|
4IP5
 
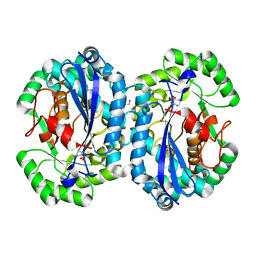 | | Crystal structure of l-fuconate dehydratase from Silicibacter sp. tm1040 liganded with Mg and d-erythronohydroxamate | | Descriptor: | (2R,3R)-N,2,3,4-TETRAHYDROXYBUTANAMIDE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Lukk, T, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-01-09 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structure of l-fuconate dehydratase from Silicibacter sp. tm1040 liganded with Mg and d-erythronohydroxamate
To be Published
|
|
4ID9
 
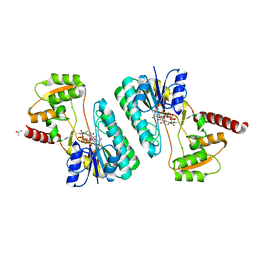 | | Crystal structure of a short-chain dehydrogenase/reductase superfamily protein from agrobacterium tumefaciens (TARGET EFI-506441) with bound nad, monoclinic form 1 | | Descriptor: | ALANINE, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Vetting, M.W, Groninger-Poe, F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-12-12 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a short-chain dehydrogenase/reductase superfamily protein from agrobacterium tumefaciens (TARGET EFI-506441) with bound nad, monoclinic form 1
To be Published
|
|
4ILK
 
 | | Crystal structure of short chain alcohol dehydrogenase (rspB) from E. coli CFT073 (EFI TARGET EFI-506413) complexed with cofactor NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, MANGANESE (II) ION, Starvation sensing protein rspB, ... | | Authors: | Lukk, T, Wichelecki, D, Imker, H.J, Gerlt, J.A, Nair, S.K. | | Deposit date: | 2012-12-31 | | Release date: | 2013-01-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Crystal structure of short chain alcohol dehydrogenase (rspB) from E. coli CFT073 (EFI TARGET EFI-506413) complexed with cofactor NADH
To be Published
|
|
4IMR
 
 | | Crystal structure of 3-oxoacyl (acyl-carrier-protein) reductase (target EFI-506442) from agrobacterium tumefaciens C58 with NADP bound | | Descriptor: | 3-oxoacyl-(Acyl-carrier-protein) reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, UNKNOWN LIGAND | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N.F, Stead, M, Love, J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-01-03 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structure of 3-Oxoacyl (Acyl-Carrier-Protein) Reductase Atu5465 from Agrobacterium Tumefaciens
To be Published
|
|
4IL0
 
 | | Crystal structure of GlucDRP from E. coli K-12 MG1655 (EFI target EFI-506058) | | Descriptor: | CITRIC ACID, GLYCEROL, Glucarate dehydratase-related protein | | Authors: | Lukk, T, Ghasempur, S, Imker, H.J, Gerlt, J.A, Nair, S.K, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-12-28 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Glucarate dehydratase and its related protein from Escherichia coli form a heterotetrameric complex.
to be published
|
|
4IJI
 
 | | Crystal structure of a glutathione transferase family member from Psuedomonas fluorescens Pf-5, target EFI-900011, with bound S-(propanoic acid)-glutathione | | Descriptor: | ACRYLIC ACID, BENZOIC ACID, Glutathione S-transferase-like protein YibF, ... | | Authors: | Vetting, M.W, Sauder, J.M, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Burley, S.K, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-12-21 | | Release date: | 2013-02-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a glutathione transferase family member from Psuedomonas fluorescens Pf-5, target EFI-900011, with bound S-(propanoic acid)-glutathione
To be Published
|
|
7RUJ
 
 | | E. coli cysteine desulfurase SufS N99A | | Descriptor: | CHLORIDE ION, Cysteine desulfurase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Dunkle, J.A, Gogar, R, Frantom, P.A. | | Deposit date: | 2021-08-17 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The beta-latch structural element of the SufS cysteine desulfurase mediates active site accessibility and SufE transpersulfurase positioning.
J.Biol.Chem., 299, 2023
|
|
7RW3
 
 | | E. coli cysteine desulfurase SufS N99D | | Descriptor: | Cysteine desulfurase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Dunkle, J.A, Gogar, R, Frantom, P.A. | | Deposit date: | 2021-08-19 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The beta-latch structural element of the SufS cysteine desulfurase mediates active site accessibility and SufE transpersulfurase positioning.
J.Biol.Chem., 299, 2023
|
|
7T6C
 
 | | E. coli dihydroorotate dehydrogenase bound to the ubiquinone surrogate DCIP | | Descriptor: | 2,6-bis(chloranyl)-4-[(4-hydroxyphenyl)amino]phenol, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Horwitz, S.M, Ambarian, J.A, Davis, K.M. | | Deposit date: | 2021-12-13 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural insights into inhibition of the drug target dihydroorotate dehydrogenase by bacterial hydroxyalkylquinolines.
Rsc Chem Biol, 3, 2022
|
|
7T5K
 
 | | E. coli dihydroorotate dehydrogenase bound to the inhibitor HQNO | | Descriptor: | 2-HEPTYL-4-HYDROXY QUINOLINE N-OXIDE, Dihydroorotate dehydrogenase (quinone), FLAVIN MONONUCLEOTIDE, ... | | Authors: | Horwitz, S.M, Ambarian, J.A, Davis, K.M. | | Deposit date: | 2021-12-12 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights into inhibition of the drug target dihydroorotate dehydrogenase by bacterial hydroxyalkylquinolines.
Rsc Chem Biol, 3, 2022
|
|
7T6H
 
 | | E. coli dihydroorotate dehydrogenase | | Descriptor: | Dihydroorotate dehydrogenase (quinone), FLAVIN MONONUCLEOTIDE, OROTIC ACID | | Authors: | Horwitz, S.M, Ambarian, J.A, Davis, K.M. | | Deposit date: | 2021-12-13 | | Release date: | 2022-03-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural insights into inhibition of the drug target dihydroorotate dehydrogenase by bacterial hydroxyalkylquinolines.
Rsc Chem Biol, 3, 2022
|
|
7T5Y
 
 | | E. coli dihydroorotate dehydrogenase bound to the inhibitor HMNQ | | Descriptor: | 3-methyl-2-[(2E)-non-2-en-1-yl]quinolin-4(1H)-one, Dihydroorotate dehydrogenase (quinone), FLAVIN MONONUCLEOTIDE, ... | | Authors: | Horwitz, S.M, Ambarian, J.A, Davis, K.M. | | Deposit date: | 2021-12-13 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural insights into inhibition of the drug target dihydroorotate dehydrogenase by bacterial hydroxyalkylquinolines.
Rsc Chem Biol, 3, 2022
|
|
7T9H
 
 | | HIV Integrase in complex with Compound-15 | | Descriptor: | (2S)-tert-butoxy[2-methyl-4-(4-methylphenyl)quinolin-3-yl]acetic acid, Integrase, MAGNESIUM ION | | Authors: | Khan, J.A, Lewis, H, Kish, K. | | Deposit date: | 2021-12-19 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Design, Synthesis, and Preclinical Profiling of GSK3739936 (BMS-986180), an Allosteric Inhibitor of HIV-1 Integrase with Broad-Spectrum Activity toward 124/125 Polymorphs.
J.Med.Chem., 65, 2022
|
|
7TGH
 
 | | Cryo-EM structure of respiratory super-complex CI+III2 from Tetrahymena thermophila | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2 iron, ... | | Authors: | Zhou, L, Maldonado, M, Padavannil, A, Guo, F, Letts, J.A. | | Deposit date: | 2022-01-07 | | Release date: | 2022-04-06 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structures of Tetrahymena 's respiratory chain reveal the diversity of eukaryotic core metabolism.
Science, 376, 2022
|
|
7T9O
 
 | | HIV Integrase in complex with Compound-25 | | Descriptor: | (2S)-tert-butoxy[4-(4,4-dimethylpiperidin-1-yl)-5-{4-[2-(4-fluorophenyl)ethoxy]phenyl}-2,6-dimethylpyridin-3-yl]acetic acid, GLYCEROL, Integrase, ... | | Authors: | Khan, J.A, Lewis, H, Kish, K. | | Deposit date: | 2021-12-19 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design, Synthesis, and Preclinical Profiling of GSK3739936 (BMS-986180), an Allosteric Inhibitor of HIV-1 Integrase with Broad-Spectrum Activity toward 124/125 Polymorphs.
J.Med.Chem., 65, 2022
|
|
7T1T
 
 | | JAK2 JH2 IN COMPLEX WITH JAK292 | | Descriptor: | (2S)-2-[({4-[(2-amino-7H-pyrrolo[2,3-d]pyrimidin-4-yl)oxy]phenyl}carbamoyl)amino]-4-phenylbutanoic acid, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Ippolito, J.A, Henry, S, Krimmer, S.G, Schlessinger, J, Jorgensen, W.L. | | Deposit date: | 2021-12-02 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Conversion of a False Virtual Screen Hit into Selective JAK2 JH2 Domain Binders Using Convergent Design Strategies
Acs Med.Chem.Lett., 13, 2022
|
|
7T0P
 
 | | JAK2 JH2 IN COMPLEX WITH JAK315 | | Descriptor: | 4'-{[5-amino-3-(4-sulfamoylanilino)-1H-1,2,4-triazole-1-carbonyl]amino}-4-(benzyloxy)[1,1'-biphenyl]-3-carboxylic acid, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Ippolito, J.A, Liosi, M.-E, Krimmer, S.G, Schlessinger, J, Jorgensen, W.L. | | Deposit date: | 2021-11-30 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Insights on JAK2 Modulation by Potent, Selective, and Cell-Permeable Pseudokinase-Domain Ligands.
J.Med.Chem., 65, 2022
|
|
7SZU
 
 | |
1TLL
 
 | | CRYSTAL STRUCTURE OF RAT NEURONAL NITRIC-OXIDE SYNTHASE REDUCTASE MODULE AT 2.3 A RESOLUTION. | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Garcin, E.D, Bruns, C.M, Lloyd, S.J, Hosfield, D.J, Tiso, M, Gachhui, R, Stuehr, D.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2004-06-09 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for isozyme-specific regulation of electron transfer in nitric-oxide synthase
J.Biol.Chem., 279, 2004
|
|
1SRR
 
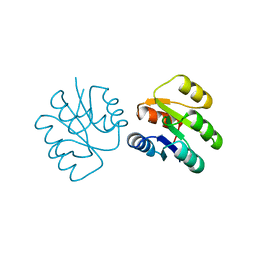 | | CRYSTAL STRUCTURE OF A PHOSPHATASE RESISTANT MUTANT OF SPORULATION RESPONSE REGULATOR SPO0F FROM BACILLUS SUBTILIS | | Descriptor: | CALCIUM ION, SPORULATION RESPONSE REGULATORY PROTEIN | | Authors: | Madhusudan, Whiteley, J.M, Hoch, J.A, Zapf, J, Xuong, N.H, Varughese, K.I. | | Deposit date: | 1996-04-10 | | Release date: | 1997-04-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a phosphatase-resistant mutant of sporulation response regulator Spo0F from Bacillus subtilis.
Structure, 4, 1996
|
|
1SZK
 
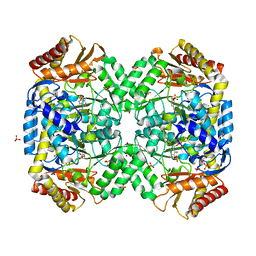 | | The structure of gamma-aminobutyrate aminotransferase mutant: E211S | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-aminobutyrate aminotransferase, ... | | Authors: | Liu, W, Peterson, P.E, Langston, J.A, Jin, X, Fisher, A.J, Toney, M.D. | | Deposit date: | 2004-04-05 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Kinetic and Crystallographic Analysis of Active Site Mutants of Escherichia coligamma-Aminobutyrate Aminotransferase.
Biochemistry, 44, 2005
|
|
1T0K
 
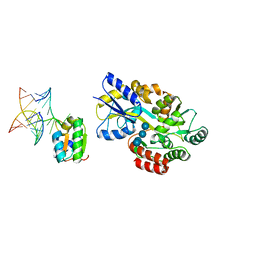 | | Joint X-ray and NMR Refinement of Yeast L30e-mRNA complex | | Descriptor: | 5'-R(*G*GP*AP*CP*GP*CP*AP*GP*AP*GP*AP*UP*GP*GP*UP*C)-3', 5'-R(*GP*AP*CP*CP*GP*GP*AP*GP*UP*GP*UP*CP*C)-3', 60S ribosomal protein L30, ... | | Authors: | Chao, J.A, Williamson, J.R. | | Deposit date: | 2004-04-09 | | Release date: | 2004-07-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Joint X-Ray and NMR Refinement of the Yeast L30e-mRNA Complex
Structure, 12, 2004
|
|
1T6I
 
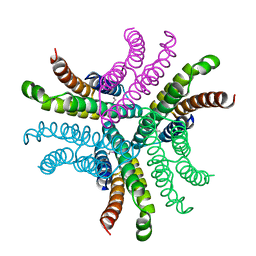 | | Nickel Superoxide Dismutase (NiSOD) Apo Structure | | Descriptor: | Superoxide dismutase [Ni] | | Authors: | Barondeau, D.P, Kassmann, C.J, Bruns, C.K, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2004-05-06 | | Release date: | 2004-07-13 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Nickel superoxide dismutase structure and mechanism.
Biochemistry, 43, 2004
|
|
