2DO7
 
 | | Solution structure of the winged helix-turn-helix motif of human CUL-4B | | Descriptor: | Cullin-4B | | Authors: | Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-27 | | Release date: | 2007-04-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the winged helix-turn-helix motif of human CUL-4B
To be Published
|
|
1IUY
 
 | |
8R5H
 
 | | Ubiquitin ligation to neosubstrate by a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-VHL-MZ1 with trapped UBE2R2~donor UB-BRD4 BD2 | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-[2-[2-[2-[2-[2-[(9~{S})-7-(4-chlorophenyl)-4,5,13-trimethyl-3-thia-1,8,11,12-tetrazatricyclo[8.3.0.0^{2,6}]trideca-2(6),4,7,10,12-pentaen-9-yl]ethanoylamino]ethoxy]ethoxy]ethoxy]ethanoylamino]-3,3-dimethyl-butanoyl]-~{N}-[[4-(4-methyl-2,3-dihydro-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, 5-azanylpentan-2-one, Bromodomain-containing protein 4, ... | | Authors: | Liwocha, J, Prabu, J.R, Kleiger, G, Schulman, B.A. | | Deposit date: | 2023-11-16 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Cullin-RING ligases employ geometrically optimized catalytic partners for substrate targeting.
Mol.Cell, 84, 2024
|
|
8CAF
 
 | | N8C_Fab3b in complex with NEDD8-CUL1(WHB) | | Descriptor: | Cullin-1, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Duda, D.M, Yanishevski, D, Henneberg, L.T, Schulman, B.A. | | Deposit date: | 2023-01-24 | | Release date: | 2023-09-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Activity-based profiling of cullin-RING E3 networks by conformation-specific probes.
Nat.Chem.Biol., 19, 2023
|
|
3RTR
 
 | | A RING E3-substrate complex poised for ubiquitin-like protein transfer: structural insights into cullin-RING ligases | | Descriptor: | Cullin-1, E3 ubiquitin-protein ligase RBX1, ZINC ION | | Authors: | Calabrese, M.F, Scott, D.C, Duda, D.M, Grace, C.R, Kurinov, I, Kriwacki, R.W, Schulman, B.A. | | Deposit date: | 2011-05-03 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | A RING E3-substrate complex poised for ubiquitin-like protein transfer: structural insights into cullin-RING ligases.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2HYE
 
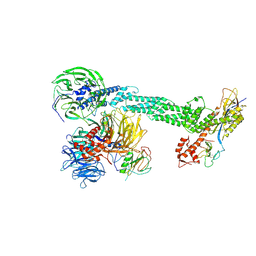 | | Crystal Structure of the DDB1-Cul4A-Rbx1-SV5V Complex | | Descriptor: | Cullin-4A, DNA damage-binding protein 1, Nonstructural protein V, ... | | Authors: | Angers, S, Li, T, Yi, X, MacCoss, M.J, Moon, R.T, Zheng, N. | | Deposit date: | 2006-08-05 | | Release date: | 2006-10-03 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery.
Nature, 443, 2006
|
|
5N4W
 
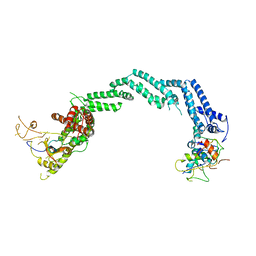 | | Crystal structure of the Cul2-Rbx1-EloBC-VHL ubiquitin ligase complex | | Descriptor: | Cullin-2, E3 ubiquitin-protein ligase RBX1, Elongin-B, ... | | Authors: | Cardote, T.A.F, Gadd, M.S, Ciulli, A. | | Deposit date: | 2017-02-11 | | Release date: | 2017-06-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Crystal Structure of the Cul2-Rbx1-EloBC-VHL Ubiquitin Ligase Complex.
Structure, 25, 2017
|
|
1U6G
 
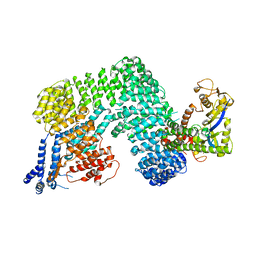 | | Crystal Structure of The Cand1-Cul1-Roc1 Complex | | Descriptor: | Cullin homolog 1, RING-box protein 1, TIP120 protein, ... | | Authors: | Goldenberg, S.J, Shumway, S.D, Cascio, T.C, Garbutt, K.C, Liu, J, Xiong, Y, Zheng, N. | | Deposit date: | 2004-07-29 | | Release date: | 2004-12-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases
Cell(Cambridge,Mass.), 119, 2004
|
|
5V89
 
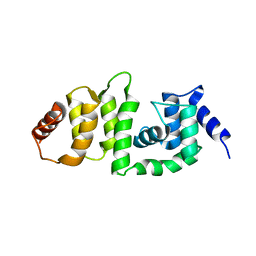 | | Structure of DCN4 PONY domain bound to CUL1 WHB | | Descriptor: | Cullin-1, DCN1-like protein 4 | | Authors: | Guy, R.K, Schulman, B.A, Scott, D.C, Hammill, J.T. | | Deposit date: | 2017-03-21 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Blocking an N-terminal acetylation-dependent protein interaction inhibits an E3 ligase.
Nat. Chem. Biol., 13, 2017
|
|
4P5O
 
 | |
7SHL
 
 | | Structure of Xenopus laevis CRL2Lrr1 (State 2) | | Descriptor: | CULLIN_2 domain-containing protein, Elongin-C, Lrr1, ... | | Authors: | Zhou, H, Brown, A. | | Deposit date: | 2021-10-09 | | Release date: | 2021-12-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of CRL2Lrr1, the E3 ubiquitin ligase that promotes DNA replication termination in vertebrates.
Nucleic Acids Res., 49, 2021
|
|
7SHK
 
 | | Structure of Xenopus laevis CRL2Lrr1 (State 1) | | Descriptor: | CULLIN_2 domain-containing protein, Elongin-C, Lrr1, ... | | Authors: | Zhou, H, Brown, A. | | Deposit date: | 2021-10-09 | | Release date: | 2021-12-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of CRL2Lrr1, the E3 ubiquitin ligase that promotes DNA replication termination in vertebrates.
Nucleic Acids Res., 49, 2021
|
|
3O2P
 
 | | A Dual E3 Mechanism for Rub1 Ligation to Cdc53: Dcn1(P)-Cdc53(WHB) | | Descriptor: | Cell division control protein 53, Defective in cullin neddylation protein 1, GLYCEROL | | Authors: | Scott, D.C, Monda, J.K, Grace, C.R.R, Duda, D.M, Kriwacki, R.W, Kurz, T, Schulman, B.A. | | Deposit date: | 2010-07-22 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.233 Å) | | Cite: | A dual E3 mechanism for Rub1 ligation to Cdc53.
Mol.Cell, 39, 2010
|
|
8RWZ
 
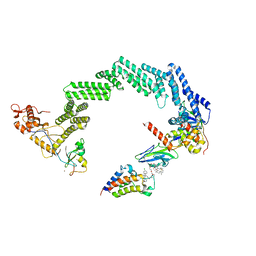 | | Open non-crosslinked structure Brd4BD2-MZ1-(NEDD8)-CRL2VHL | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-[2-[2-[2-[2-[2-[(9~{S})-7-(4-chlorophenyl)-4,5,13-trimethyl-3-thia-1,8,11,12-tetrazatricyclo[8.3.0.0^{2,6}]trideca-2(6),4,7,10,12-pentaen-9-yl]ethanoylamino]ethoxy]ethoxy]ethoxy]ethanoylamino]-3,3-dimethyl-butanoyl]-~{N}-[[4-(4-methyl-2,3-dihydro-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Bromodomain-containing protein 4, Cullin-2, ... | | Authors: | Ciulli, A, Crowe, C, Nakacone, M.A. | | Deposit date: | 2024-02-05 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Open non-crosslinked structure Brd4BD2-MZ1-(NEDD8)-CRL2VHL
To Be Published
|
|
8RX0
 
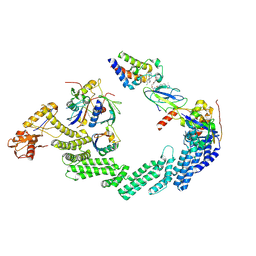 | | (NEDD8)-CRL2VHL-MZ1-Brd4BD2-Ub(G76S, K48C)-UBE2R1(C93K, S138C, C191S, C223S)-Ub | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-[2-[2-[2-[2-[2-[(9~{S})-7-(4-chlorophenyl)-4,5,13-trimethyl-3-thia-1,8,11,12-tetrazatricyclo[8.3.0.0^{2,6}]trideca-2(6),4,7,10,12-pentaen-9-yl]ethanoylamino]ethoxy]ethoxy]ethoxy]ethanoylamino]-3,3-dimethyl-butanoyl]-~{N}-[[4-(4-methyl-2,3-dihydro-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Crowe, C, Nakasone, M.A, Ciulli, A. | | Deposit date: | 2024-02-05 | | Release date: | 2024-03-06 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | (NEDD8)-CRL2VHL-MZ1-Brd4BD2-Ub(G76S, K48C)-UBE2R1(C93K, S138C, C191S, C223S)-Ub
To Be Published
|
|
6TTU
 
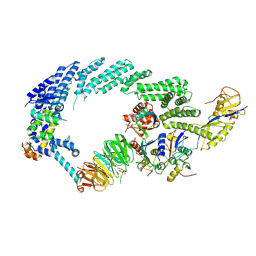 | | Ubiquitin Ligation to substrate by a cullin-RING E3 ligase at 3.7A resolution: NEDD8-CUL1-RBX1 N98R-SKP1-monomeric b-TRCP1dD-IkBa-UB~UBE2D2 | | Descriptor: | CYS-LYS-LYS-ALA-ARG-HIS-ASP-SEP-GLY, Cullin-1, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Baek, K, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2019-12-30 | | Release date: | 2020-02-12 | | Last modified: | 2020-03-04 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | NEDD8 nucleates a multivalent cullin-RING-UBE2D ubiquitin ligation assembly.
Nature, 578, 2020
|
|
3DQV
 
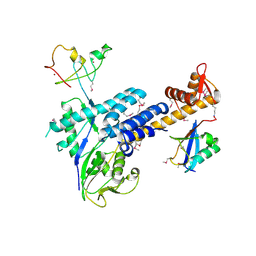 | | Structural Insights into NEDD8 Activation of Cullin-RING Ligases: Conformational Control of Conjugation | | Descriptor: | Cullin-5, NEDD8, Rbx1, ... | | Authors: | Duda, D.M, Borg, L.A, Scott, D.C, Hunt, H.W, Hammel, M, Schulman, B.A. | | Deposit date: | 2008-07-09 | | Release date: | 2008-09-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation.
Cell(Cambridge,Mass.), 134, 2008
|
|
3TDZ
 
 | | N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex: Structure of a human Cul1WHB-Dcn1P-stapled acetylated Ubc12N complex | | Descriptor: | Cullin-1, DCN1-like protein 1, STAPLED PEPTIDE | | Authors: | Scott, D.C, Monda, J.K, Bennett, E.J, Harper, J.W, Schulman, B.A. | | Deposit date: | 2011-08-11 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex.
Science, 334, 2011
|
|
3TDU
 
 | | N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex: Structure of a human Cul1WHB-Dcn1P-acetylated Ubc12N complex | | Descriptor: | Cullin-1, DCN1-like protein 1, NEDD8-conjugating enzyme Ubc12 | | Authors: | Scott, D.C, Monda, J.K, Bennett, E.J, Harper, J.W, Schulman, B.A. | | Deposit date: | 2011-08-11 | | Release date: | 2011-10-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex.
Science, 334, 2011
|
|
3DPL
 
 | |
6V9I
 
 | | cryo-EM structure of Cullin5 bound to RING-box protein 2 (Cul5-Rbx2) | | Descriptor: | Immunoglobulin G-binding protein G,Cullin-5, RING-box protein 2, ZINC ION | | Authors: | Komives, E.A, Lumpkin, R.J, Baker, R.W, Leschziner, A.E. | | Deposit date: | 2019-12-13 | | Release date: | 2020-04-29 | | Last modified: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Structure and dynamics of the ASB9 CUL-RING E3 Ligase.
Nat Commun, 11, 2020
|
|
3O6B
 
 | | A Dual E3 Mechanism for Rub1 Ligation to Cdc53: Dcn1(P)-Cdc53(WHB) low resolution | | Descriptor: | Cell division control protein 53, Defective in cullin neddylation protein 1 | | Authors: | Scott, D.C, Monda, J.K, Grace, C.R.R, Duda, D.M, Kriwacki, R.W, Kurz, T, Schulman, B.A. | | Deposit date: | 2010-07-28 | | Release date: | 2010-09-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A dual E3 mechanism for Rub1 ligation to Cdc53.
Mol.Cell, 39, 2010
|
|
8B3G
 
 | | C(N)RL4CSA-UVSSA-E2-ubiquitin complex. | | Descriptor: | Cullin-4A, DNA damage-binding protein 1, DNA excision repair protein ERCC-8, ... | | Authors: | Kokic, G, Cramer, P. | | Deposit date: | 2022-09-16 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural basis for RNA polymerase II ubiquitylation and inactivation in transcription-coupled repair.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8CDK
 
 | |
8CDJ
 
 | | CAND1 b-hairpin++-SCF-SKP2 CAND1 rolling SCF engaged | | Descriptor: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Baek, K, Schulman, B.A. | | Deposit date: | 2023-01-31 | | Release date: | 2023-04-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Systemwide disassembly and assembly of SCF ubiquitin ligase complexes.
Cell, 186, 2023
|
|
