8VR2
 
 | | Crystal structure of the Pcryo_0617 oxidoreductase/decarboxylase from Psychrobacter cryohalolentis K5 in the presence of NAD and UDP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, NAD-dependent epimerase/dehydratase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Bockhaus, N.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2024-01-20 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biochemical Investigation of the Enzymes Required for the Production of 2,3,4-triacetoamido-2,3,4-trideoxy-l-arabinose in Psychrobacter cryohalolentis K5
To Be Published
|
|
8V4H
 
 | | X-ray structure of the NADP-dependent reductase from Campylobacter jejuni responsible for the synthesis of CDP-glucitol in the presence of CDP-glucitol | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Putative nucleotide sugar dehydratase, ... | | Authors: | Thoden, J.B, Schumann, M.E, Holden, H.M, Raushel, F.M. | | Deposit date: | 2023-11-29 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biosynthesis of Cytidine Diphosphate-6-d-Glucitol for the Capsular Polysaccharides of Campylobacter jejuni.
Biochemistry, 63, 2024
|
|
8V4G
 
 | | X-ray structure of the NADP-dependent reductase from Campylobacter jejuni responsible for the synthesis of CDP-glucitol in the presence of CDP and NADP | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CYTIDINE-5'-DIPHOSPHATE, ... | | Authors: | Schumann, M.E, Thoden, J.B, Holden, H.M, Raushel, F.M. | | Deposit date: | 2023-11-29 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biosynthesis of Cytidine Diphosphate-6-d-Glucitol for the Capsular Polysaccharides of Campylobacter jejuni.
Biochemistry, 63, 2024
|
|
8JQK
 
 | | Crystal structure of a carbonyl reductase SSCR mutant from Sporobolomyces Salmonicolor | | Descriptor: | Aldehyde reductase 2 | | Authors: | Zhang, H.L, Li, Q, Liu, W.D, Chen, X, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2023-06-14 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Engineering a Carbonyl Reductase to Simultaneously Increase Activity Toward Bulky Ketone and Isopropanol for Dynamic Kinetic Asymmetric Reduction via Enzymatic Hydrogen Transfer
Acs Catalysis, 13, 2023
|
|
8JQJ
 
 | | Crystal structure of carbonyl reductase SSCR mutant 1 from Sporobolomyces Salmonicolor | | Descriptor: | Aldehyde reductase 2 | | Authors: | Zhang, H.L, Li, Q, Liu, W.D, Chen, X, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2023-06-14 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Engineering a Carbonyl Reductase to Simultaneously Increase Activity Toward Bulky Ketone and Isopropanol for Dynamic Kinetic Asymmetric Reduction via Enzymatic Hydrogen Transfer
Acs Catalysis, 13, 2023
|
|
8H61
 
 | | Ketoreductase CpKR mutant - M2 | | Descriptor: | Mutant M2 of ketoreductase CpKR, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Chen, C, Pan, J, Xu, J.H. | | Deposit date: | 2022-10-14 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Computational redesign of a robust ketoreductase for asymmetric synthesis of enantiopure diltiazem precursor.
To Be Published
|
|
8GZU
 
 | | Cryo-EM structure of Tetrahymena thermophila respiratory Megacomplex MC (IV2+I+III2+II)2 | | Descriptor: | 2 iron, 2 sulfur cluster-binding protein, 2-oxoglutarate/malate carrier protein, ... | | Authors: | Wu, M.C, Hu, Y.Q, Han, F.Z, Zhou, L. | | Deposit date: | 2022-09-27 | | Release date: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Structures of Tetrahymena thermophila respiratory megacomplexes on the tubular mitochondrial cristae.
Nat Commun, 14, 2023
|
|
8GYM
 
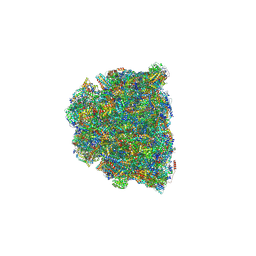 | | Cryo-EM structure of Tetrahymena thermophila respiratory mega-complex MC IV2+(I+III2+II)2 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ... | | Authors: | Wu, M.C, Hu, Y.Q, Han, F.Z, Zhou, L. | | Deposit date: | 2022-09-23 | | Release date: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structures of Tetrahymena thermophila respiratory megacomplexes on the tubular mitochondrial cristae.
Nat Commun, 14, 2023
|
|
8GJH
 
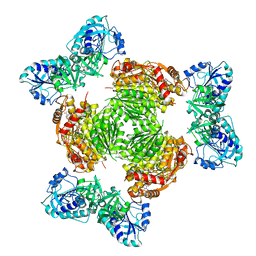 | | Salmonella ArnA | | Descriptor: | Bifunctional polymyxin resistance protein ArnA, URIDINE-5'-DIPHOSPHATE-GLUCURONIC ACID | | Authors: | Sousa, M.C, Mitchell, M.E, Gatzeva-Topalova, P.Z. | | Deposit date: | 2023-03-15 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Targeting the Conformational Change in ArnA Dehydrogenase for Selective Inhibition of Polymyxin Resistance.
Biochemistry, 62, 2023
|
|
8GJB
 
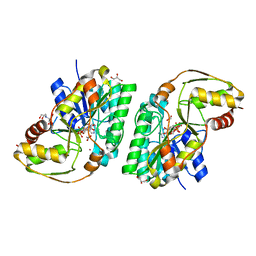 | | L-threonine 3-Dehydrogenase from Trypanosoma cruzi in complex with NAD and acetate | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Mercaldi, G.F, Faria, J.N, Fagundes, M, Bezerra, E.H.S, Cordeiro, A.T. | | Deposit date: | 2023-03-15 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure, allosteric regulation and metabolic activity of L-threonine 3-Dehydrogenase from Trypanosoma cruzi
To Be Published
|
|
8GIL
 
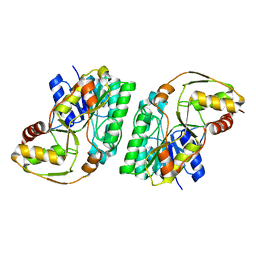 | | L-threonine 3-Dehydrogenase from Trypanosoma cruzi (apo form) | | Descriptor: | L-threonine 3-dehydrogenase, POTASSIUM ION | | Authors: | Faria, J.N, Mercaldi, G.F, Fagundes, M, Bezerra, E.H.S, Cordeiro, A.T. | | Deposit date: | 2023-03-14 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure, allosteric regulation and metabolic activity of L-threonine 3-Dehydrogenase from Trypanosoma cruzi
To Be Published
|
|
8FTN
 
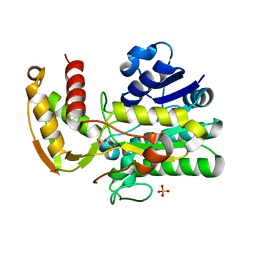 | | E. coli ArnA dehydrogenase domain mutant - N492A | | Descriptor: | Bifunctional UDP-4-amino-4-deoxy-L-arabinose formyltransferase/UDP-glucuronic acid oxidase ArnA, SULFATE ION | | Authors: | Sousa, M.C, Mitchell, M.E. | | Deposit date: | 2023-01-12 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Targeting the Conformational Change in ArnA Dehydrogenase for Selective Inhibition of Polymyxin Resistance.
Biochemistry, 62, 2023
|
|
8FIP
 
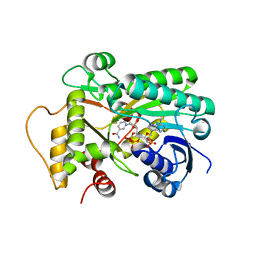 | | Hypothetical anthocyanidin reducatase from Sorghum bicolor- NADP+ complex | | Descriptor: | Epimerase domain-containing protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, B, Kang, C. | | Deposit date: | 2022-12-16 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Similarities and Overlapping Activities among Dihydroflavonol 4-Reductase, Flavanone 4-Reductase, and Anthocyanidin Reductase Offer Metabolic Flexibility in the Flavonoid Pathway.
Int J Mol Sci, 24, 2023
|
|
8FIO
 
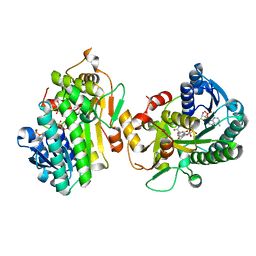 | | Hypothetical anthocyanidin reductase from Sorghum bicolor-NADP(H) and naringenin complex | | Descriptor: | Epimerase domain-containing protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NARINGENIN | | Authors: | Zhang, B, Kang, C. | | Deposit date: | 2022-12-16 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Similarities and Overlapping Activities among Dihydroflavonol 4-Reductase, Flavanone 4-Reductase, and Anthocyanidin Reductase Offer Metabolic Flexibility in the Flavonoid Pathway.
Int J Mol Sci, 24, 2023
|
|
8FEW
 
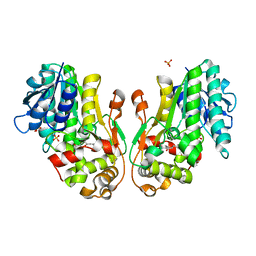 | | Flavanone 4-Reductase from Sorghum bicolor-naringenin complex | | Descriptor: | 3-deoxyanthocyanidin synthase, NARINGENIN, SULFATE ION | | Authors: | Zhang, B, Kang, C. | | Deposit date: | 2022-12-06 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural Similarities and Overlapping Activities among Dihydroflavonol 4-Reductase, Flavanone 4-Reductase, and Anthocyanidin Reductase Offer Metabolic Flexibility in the Flavonoid Pathway.
Int J Mol Sci, 24, 2023
|
|
8FEV
 
 | | Flavanone 4-Reductase from Sorghum bicolor-NADP(H) and dihydroquercetin complex | | Descriptor: | (2R,3R)-2-(3,4-DIHYDROXYPHENYL)-3,5,7-TRIHYDROXY-2,3-DIHYDRO-4H-CHROMEN-4-ONE, 3-deoxyanthocyanidin synthase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Zhang, B, Kang, C. | | Deposit date: | 2022-12-06 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural Similarities and Overlapping Activities among Dihydroflavonol 4-Reductase, Flavanone 4-Reductase, and Anthocyanidin Reductase Offer Metabolic Flexibility in the Flavonoid Pathway.
Int J Mol Sci, 24, 2023
|
|
8FEU
 
 | | Flavanone 4-Reductase from Sorghum bicolor-NADP(H) and naringenin complex | | Descriptor: | 3-deoxyanthocyanidin synthase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NARINGENIN, ... | | Authors: | Zhang, B, Kang, C. | | Deposit date: | 2022-12-06 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural Similarities and Overlapping Activities among Dihydroflavonol 4-Reductase, Flavanone 4-Reductase, and Anthocyanidin Reductase Offer Metabolic Flexibility in the Flavonoid Pathway.
Int J Mol Sci, 24, 2023
|
|
8FET
 
 | | Flavanone 4-Reductase from Sorghum bicolor-NADP(H) complex | | Descriptor: | 3-deoxyanthocyanidin synthase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, B, Kang, C. | | Deposit date: | 2022-12-06 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Structural Similarities and Overlapping Activities among Dihydroflavonol 4-Reductase, Flavanone 4-Reductase, and Anthocyanidin Reductase Offer Metabolic Flexibility in the Flavonoid Pathway.
Int J Mol Sci, 24, 2023
|
|
8FEN
 
 | | Panicum vigratum Dihydroflavonol 4-reductase complexed with NADP and DHQ | | Descriptor: | (2R,3R)-2-(3,4-DIHYDROXYPHENYL)-3,5,7-TRIHYDROXY-2,3-DIHYDRO-4H-CHROMEN-4-ONE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Panicum virgatum Dihydroflavonol 4-Reductase | | Authors: | Lewis, J.A, Kang, C. | | Deposit date: | 2022-12-06 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Similarities and Overlapping Activities among Dihydroflavonol 4-Reductase, Flavanone 4-Reductase, and Anthocyanidin Reductase Offer Metabolic Flexibility in the Flavonoid Pathway.
Int J Mol Sci, 24, 2023
|
|
8FEM
 
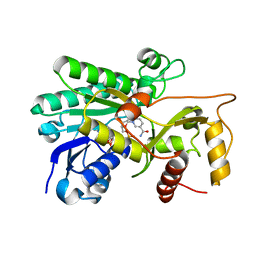 | | Panicum vigratum Dihydroflavonol 4-reductase complexed with NADP | | Descriptor: | Dihydroflavonol 4-Reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Lewis, J.A, Kang, C. | | Deposit date: | 2022-12-06 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural Similarities and Overlapping Activities among Dihydroflavonol 4-Reductase, Flavanone 4-Reductase, and Anthocyanidin Reductase Offer Metabolic Flexibility in the Flavonoid Pathway.
Int J Mol Sci, 24, 2023
|
|
8EWU
 
 | | X-ray structure of the GDP-6-deoxy-4-keto-D-lyxo-heptose-4-reductase from Campylobacter jejuni HS:15 | | Descriptor: | 1,2-ETHANEDIOL, GDP-L-fucose synthase, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Thoden, J.B, Xiang, D.F, Ghosh, M.K, Riegert, A.S, Raushel, F.M, Holden, H.M. | | Deposit date: | 2022-10-24 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Bifunctional Epimerase/Reductase Enzymes Facilitate the Modulation of 6-Deoxy-Heptoses Found in the Capsular Polysaccharides of Campylobacter jejuni.
Biochemistry, 62, 2023
|
|
8ESZ
 
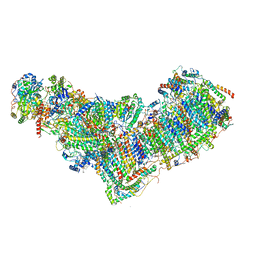 | | Structure of mitochondrial complex I from Drosophila melanogaster, Helix-locked state | | Descriptor: | (2R)-3-{[(S)-hydroxy(3-methylbutoxy)phosphoryl]oxy}-2-(octanoyloxy)propyl decanoate, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ... | | Authors: | Padavannil, A, Letts, J.A. | | Deposit date: | 2022-10-15 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Resting mitochondrial complex I from Drosophila melanogaster adopts a helix-locked state.
Elife, 12, 2023
|
|
8ESW
 
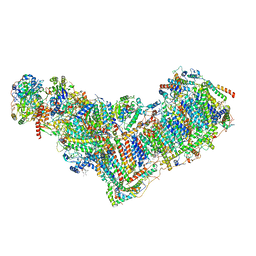 | | Structure of mitochondrial complex I from Drosophila melanogaster, Flexible-class 1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Padavannil, A, Letts, J.A. | | Deposit date: | 2022-10-15 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Resting mitochondrial complex I from Drosophila melanogaster adopts a helix-locked state.
Elife, 12, 2023
|
|
8E73
 
 | | Vigna radiata supercomplex I+III2 (full bridge) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CARDIOLIPIN, ... | | Authors: | Maldonado, M, Letts, J.A. | | Deposit date: | 2022-08-23 | | Release date: | 2023-01-11 | | Last modified: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Plant-specific features of respiratory supercomplex I + III 2 from Vigna radiata.
Nat.Plants, 9, 2023
|
|
8DU0
 
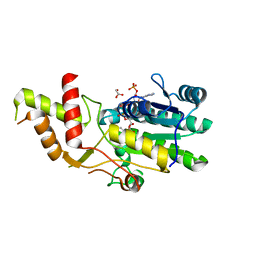 | |
