4XLT
 
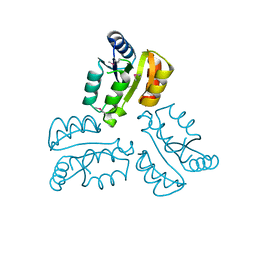 | | Crystal structure of response regulator receiver protein from Dyadobacter fermentans DSM 18053 | | Descriptor: | Response regulator receiver protein | | Authors: | Chang, C, Cuff, M, Holowicki, J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-01-13 | | Release date: | 2015-01-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of response regulator receiver protein from Dyadobacter fermentans DSM 18053
To Be Published
|
|
1HEY
 
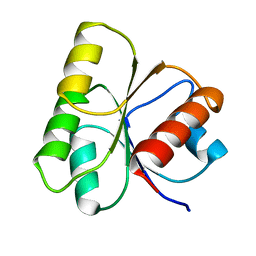 | |
4D6Y
 
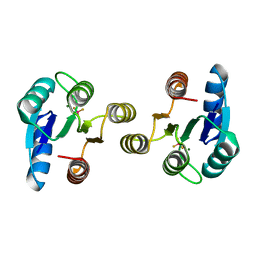 | | Crystal structure of the receiver domain of NtrX from Brucella abortus in complex with beryllofluoride and magnesium | | Descriptor: | BACTERIAL REGULATORY, FIS FAMILY PROTEIN, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Otero, L.H, Fernandez, I, Carrica, M.C, Klinke, S, Goldbaum, F.A. | | Deposit date: | 2014-11-18 | | Release date: | 2015-07-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Snapshots of Conformational Changes Shed Light Into the Ntrx Receiver Domain Signal Transduction Mechanism
J.Mol.Biol., 427, 2015
|
|
1QMP
 
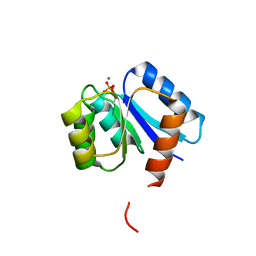 | | Phosphorylated aspartate in the crystal structure of the sporulation response regulator, Spo0A | | Descriptor: | CALCIUM ION, Stage 0 sporulation protein A | | Authors: | Lewis, R.J, Brannigan, J.A, Muchova, K, Barak, I, Wilkinson, A.J. | | Deposit date: | 1999-10-04 | | Release date: | 1999-11-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphorylated aspartate in the structure of a response regulator protein.
J. Mol. Biol., 294, 1999
|
|
8FK2
 
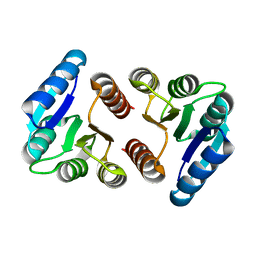 | | The N-terminal VicR from Streptococcus mutans | | Descriptor: | Putative response regulator CovR VicR-like protein | | Authors: | Zhang, H, Wu, H. | | Deposit date: | 2022-12-20 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Small Molecule Attenuates Bacterial Virulence by Targeting Conserved Response Regulator.
Mbio, 14, 2023
|
|
6EBB
 
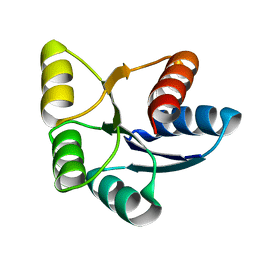 | |
6EXR
 
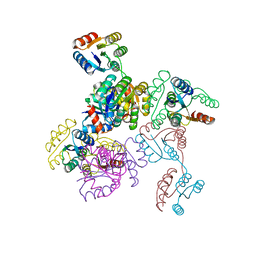 | | CHEMOTAXIS PROTEIN CHEY FROM Pyrococcus horikoshiI | | Descriptor: | 120aa long hypothetical chemotaxis protein (CheY) | | Authors: | Paithankar, K.S, Enderle, M.E, Wirthensohn, D, Grininger, M, Oesterhelt, D. | | Deposit date: | 2017-11-09 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structure of the archaeal chemotaxis protein CheY in a domain-swapped dimeric conformation.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
8GC6
 
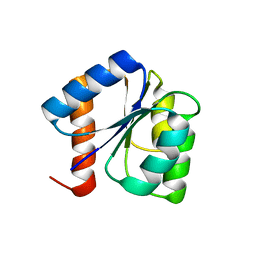 | |
6OEC
 
 | | Yeast Spc42 Trimeric Coiled-Coil Amino Acids 181-211 fused to PDB: 3H5I | | Descriptor: | CALCIUM ION, Response regulator/sensory box protein/GGDEF domain protein,Spindle pole body component SPC42 | | Authors: | Drennan, A.C, Shivaani, K, Seeger, M.A, Andreas, M.P, Gardner, J.M, Sether, E.K.R, Jasperson, S.L, Rayment, I. | | Deposit date: | 2019-03-27 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.514 Å) | | Cite: | Structure and function of Spc42 coiled-coils in yeast centrosome assembly and duplication.
Mol.Biol.Cell, 30, 2019
|
|
6ONT
 
 | | Structure of the Francisella response regulator 1452 receiver domain | | Descriptor: | CALCIUM ION, CHLORIDE ION, SODIUM ION, ... | | Authors: | Milton, M.E, Cavanagh, J. | | Deposit date: | 2019-04-22 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Francisella novicidaTwo-Component System Response Regulator BfpR Modulates iglC Gene Expression, Antimicrobial Peptide Resistance, and Biofilm Production.
Front Cell Infect Microbiol, 10, 2020
|
|
8IMW
 
 | |
7W9H
 
 | | Crystal structure of the receiver domain of the transcription regulator FleR from Pseudomonas aeruginosa | | Descriptor: | ACETATE ION, CALCIUM ION, Response regulator protein FleR | | Authors: | Sahoo, P.K, Sheenu, n, Jain, D. | | Deposit date: | 2021-12-09 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | REC domain stabilizes the active heptamer of sigma 54 -dependent transcription factor, FleR from Pseudomonas aeruginosa.
Iscience, 26, 2023
|
|
4LE0
 
 | | Crystal structure of the receiver domain of DesR in complex with beryllofluoride and magnesium | | Descriptor: | ACETATE ION, BERYLLIUM TRIFLUORIDE ION, GLYCEROL, ... | | Authors: | Trajtenberg, F, Larrieux, N, Buschiazzo, A. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Allosteric activation of bacterial response regulators: the role of the cognate histidine kinase beyond phosphorylation.
MBio, 5, 2014
|
|
6IS1
 
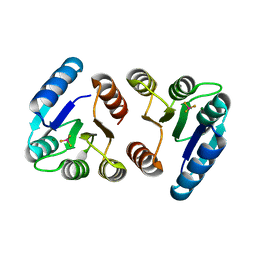 | |
3KCN
 
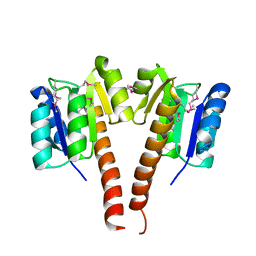 | |
5X5J
 
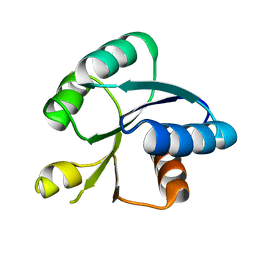 | | Crystal structure of response regulator AdeR receiver domain | | Descriptor: | AdeR | | Authors: | Wen, Y. | | Deposit date: | 2017-02-16 | | Release date: | 2017-08-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Mechanistic insight into how multidrug resistant Acinetobacter baumannii response regulator AdeR recognizes an intercistronic region.
Nucleic Acids Res., 45, 2017
|
|
3KHT
 
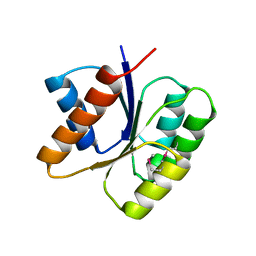 | |
6IS2
 
 | |
4LX8
 
 | |
6IS3
 
 | |
5N2N
 
 | | Crystal structure of the receiver domain of the histidine kinase CKI1 from Arabidopsis thaliana complexed with Mg2+ and BeF3- | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Histidine kinase CKI1, MAGNESIUM ION | | Authors: | Otrusinova, O, Demo, G, Padrta, P, Jasenakova, Z, Pekarova, B, Gelova, Z, Szmitkowska, A, Kaderavek, P, Jansen, S, Zachrdla, M, Klumpler, T, Marek, J, Hritz, J, Janda, L, Iwai, H, Wimmerova, M, Hejatko, J, Zidek, L. | | Deposit date: | 2017-02-08 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Conformational dynamics are a key factor in signaling mediated by the receiver domain of a sensor histidine kinase from Arabidopsis thaliana.
J. Biol. Chem., 292, 2017
|
|
6IFH
 
 | | Unphosphorylated Spo0F from Paenisporosarcina sp. TG-14 | | Descriptor: | MAGNESIUM ION, Sporulation initiation phosphotransferase F | | Authors: | Lee, J.H, Lee, C.W. | | Deposit date: | 2018-09-20 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of unphosphorylated Spo0F from Paenisporosarcina sp. TG-14, a psychrophilic bacterium isolated from an Antarctic glacier
Biodesign, 6(4), 2019
|
|
6XVU
 
 | |
5XJP
 
 | | Crystal structure of response regulator AdeR receiver domain with Mg | | Descriptor: | AdeR, MAGNESIUM ION | | Authors: | Wen, Y. | | Deposit date: | 2017-05-03 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.597 Å) | | Cite: | Mechanistic insight into how multidrug resistant Acinetobacter baumannii response regulator AdeR recognizes an intercistronic region.
Nucleic Acids Res., 45, 2017
|
|
6M8O
 
 | |
