4IYS
 
 | | Structural and ligand binding properties of the Bateman domain of human magnesium transporters CNNM2 and CNNM4 | | Descriptor: | Metal transporter CNNM2 | | Authors: | Corral-Rodriguez, M.A, Stuiver, M, Encinar, J.A, Spiwok, V, Gomez-Garcia, I, Oyenarte, I, Ereno-Orbea, J, Terashima, H, Accardi, A, Diercks, T, Muller, D, Martinez-Cruz, L.A. | | Deposit date: | 2013-01-29 | | Release date: | 2014-03-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and ligand binding properties of the Bateman domain of human magnesium transporters CNNM2 and CNNM4
To be Published
|
|
4IY0
 
 | | Structural and ligand binding properties of the Bateman domain of human magnesium transporters CNNM2 and CNNM4 | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, Metal transporter CNNM2 | | Authors: | Corral-Rodriguez, M.A, Stuiver, M, Encinar, J.A, Spiwok, V, Gomez-Garcia, I, Oyenarte, I, Ereno-Orbea, J, Terashima, H, Accardi, A, Diercks, T, Muller, D, Martinez-Cruz, L.A. | | Deposit date: | 2013-01-28 | | Release date: | 2014-03-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and ligand binding properties of the Bateman domain of human magnesium transporters CNNM2 and CNNM4
To be Published
|
|
6UOF
 
 | | 1.2 Angstrom Resolution Crystal Structure of CBS Domains of Transcriptional Regulator from Streptococcus pneumoniae | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-10-14 | | Release date: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | 1.2 Angstrom Resolution Crystal Structure of CBS Domains of Transcriptional Regulator from Streptococcus pneumoniae
To Be Published
|
|
3FWR
 
 | |
3FWS
 
 | | Crystal Structure of the CBS domains from the Bacillus subtilis CcpN repressor complexed with AppNp, phosphate and magnesium ions | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Chaix, D, Arold, S, Hoh, F, Declerck, N. | | Deposit date: | 2009-01-19 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Ligand recognition by the energy sensor domain of the CcpN repressor
To be Published
|
|
3GBY
 
 | | Crystal structure of a protein with unknown function CT1051 from Chlorobium tepidum | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SULFATE ION, Uncharacterized protein CT1051 | | Authors: | Fan, Y, Chang, C, Sather, A, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-20 | | Release date: | 2009-03-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of a protein with unknown function CT1051 from Chlorobium tepidum
To be Published
|
|
3FV6
 
 | |
3GHD
 
 | | Crystal structure of a cystathionine beta-synthase domain protein fused to a Zn-ribbon-like domain | | Descriptor: | a cystathionine beta-synthase domain protein fused to a Zn-ribbon-like domain | | Authors: | Dong, A, Xu, X, Chruszcz, M, Brown, G, Proudfoot, M, Edwards, A.M, Joachimiak, A, Minor, W, Savchenko, A, Yaleunin, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-03-03 | | Release date: | 2009-03-31 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of a cystathionine beta-synthase domain protein fused to a Zn-ribbon-like domain
To be Published
|
|
3HF7
 
 | | The Crystal Structure of a CBS-domain Pair with Bound AMP from Klebsiella pneumoniae to 2.75A | | Descriptor: | ADENOSINE MONOPHOSPHATE, uncharacterized CBS-domain protein | | Authors: | Stein, A.J, Nocek, B, Wu, R, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-11 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Crystal Structure of a CBS-domain Pair with Bound AMP from Klebsiella pneumoniae to 2.75A
To be Published
|
|
3I8N
 
 | |
3JTF
 
 | | The CBS Domain Pair Structure of a magnesium and cobalt efflux protein from Bordetella parapertussis in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Magnesium and cobalt efflux protein, SULFATE ION | | Authors: | Cuff, M.E, Tesar, C, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-11 | | Release date: | 2009-12-22 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The CBS Domain Pair Structure of a magnesium and cobalt efflux protein from Bordetella parapertussis in complex with AMP
TO BE PUBLISHED
|
|
3K2V
 
 | | Structure of the CBS pair of a putative D-arabinose 5-phosphate isomerase from Klebsiella pneumoniae subsp. pneumoniae. | | Descriptor: | CYTIDINE 5'-MONOPHOSPHATE 3-DEOXY-BETA-D-GULO-OCT-2-ULO-PYRANOSONIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Cuff, M.E, Volkart, L, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-30 | | Release date: | 2009-12-22 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the CBS pair of a putative D-arabinose 5-phosphate isomerase from Klebsiella pneumoniae subsp. pneumoniae.
TO BE PUBLISHED
|
|
3KH5
 
 | | Crystal Structure of Protein MJ1225 from Methanocaldococcus jannaschii, a putative archaeal homolog of g-AMPK. | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, SULFATE ION, ... | | Authors: | Gomez Garcia, I, Oyenarte, I, Martinez-Cruz, L.A. | | Deposit date: | 2009-10-30 | | Release date: | 2010-04-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of protein MJ1225 from Methanocaldococcus jannaschii shows strong conservation of key structural features seen in the eukaryal gamma-AMPK.
J.Mol.Biol., 65, 2010
|
|
3KPC
 
 | | Crystal Structure of the CBS domain pair of protein MJ0100 in complex with 5 -methylthioadenosine and S-adenosyl-L-methionine | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, S-ADENOSYLMETHIONINE, Uncharacterized protein MJ0100 | | Authors: | Lucas, M, Oyenarte, I, Garcia, I.G, Arribas, E.A, Encinar, J.A, Kortazar, D, Fernandez, J.A, Mato, J.M, Martinez-Chantar, M.L, Martinez-Cruz, L.A. | | Deposit date: | 2009-11-16 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Binding of S-Methyl-5'-Thioadenosine and S-Adenosyl-l-Methionine to Protein MJ0100 Triggers an Open-to-Closed Conformational Change in Its CBS Motif Pair.
J.Mol.Biol., 396, 2010
|
|
3KPD
 
 | | Crystal Structure of the CBS domain pair of protein MJ0100 in complex with 5 -methylthioadenosine and S-adenosyl-L-methionine. | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, S-ADENOSYLMETHIONINE, Uncharacterized protein MJ0100 | | Authors: | Lucas, M, Oyenarte, I, Garcia, I.G, Arribas, E.A, Encinar, J.A, Kortazar, D, Fernandez, J.A, Mato, J.M, Martinez-Chantar, M.L, Martinez-Cruz, L.A. | | Deposit date: | 2009-11-16 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Binding of S-Methyl-5'-Thioadenosine and S-Adenosyl-l-Methionine to Protein MJ0100 Triggers an Open-to-Closed Conformational Change in Its CBS Motif Pair.
J.Mol.Biol., 396, 2010
|
|
3KPB
 
 | | Crystal Structure of the CBS domain pair of protein MJ0100 in complex with 5 -methylthioadenosine and S-adenosyl-L-methionine. | | Descriptor: | GLYCEROL, S-ADENOSYLMETHIONINE, Uncharacterized protein MJ0100 | | Authors: | Lucas, M, Oyenarte, I, Garcia, I.G, Arribas, E.A, Encinar, J.A, Kortazar, D, Fernandez, J.A, Mato, J.M, Martinez-Cruz, L.A. | | Deposit date: | 2009-11-16 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Binding of S-Methyl-5'-Thioadenosine and S-Adenosyl-l-Methionine to Protein MJ0100 Triggers an Open-to-Closed Conformational Change in Its CBS Motif Pair.
J.Mol.Biol., 396, 2010
|
|
3LFZ
 
 | | Crystal Structure of Protein MJ1225 from Methanocaldococcus jannaschii, a putative archaeal homolog of g-AMPK. | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gomez-Garcia, I, Oyenarte, I, Kortazar, D, Martinez-Cruz, L.A. | | Deposit date: | 2010-01-19 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of protein MJ1225 from Methanocaldococcus jannaschii shows strong conservation of key structural features seen in the eukaryal gamma-AMPK.
J.Mol.Biol., 65, 2010
|
|
3LHH
 
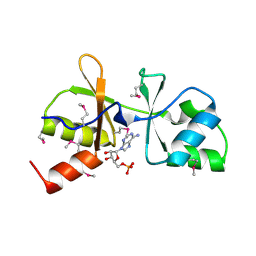 | | The crystal structure of CBS domain protein from Shewanella oneidensis MR-1. | | Descriptor: | ADENOSINE MONOPHOSPHATE, CBS domain protein | | Authors: | Tan, K, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-22 | | Release date: | 2010-02-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of CBS domain protein from Shewanella oneidensis MR-1.
To be Published
|
|
3LQN
 
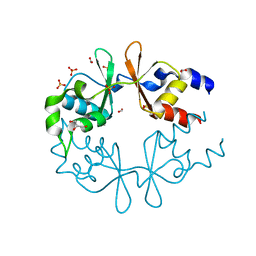 | | Crystal Structure of CBS Domain-containing Protein of Unknown Function from Bacillus anthracis str. Ames Ancestor | | Descriptor: | CBS domain protein, FORMIC ACID, GLYCEROL, ... | | Authors: | Kim, Y, Mulligan, R, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-02-09 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of CBS Domain-containing Protein of Unknown Function from Bacillus anthracis str. Ames Ancestor
To be Published
|
|
3LFR
 
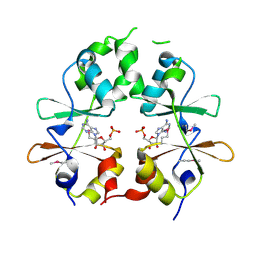 | |
2O16
 
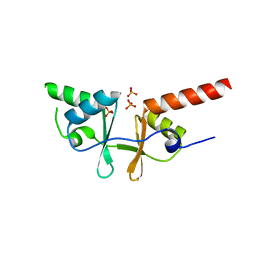 | | Crystal structure of a putative acetoin utilization protein (AcuB) from Vibrio cholerae | | Descriptor: | Acetoin utilization protein AcuB, putative, PHOSPHATE ION | | Authors: | Patskovsky, Y, Bonanno, J.B, Rutter, M, Bain, K.T, Powell, A, Slocombe, A, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-28 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a putative acetoin utilization protein (AcuB) from Vibrio cholerae
To be Published
|
|
2NYE
 
 | | Crystal structure of the Bateman2 domain of yeast Snf4 | | Descriptor: | Nuclear protein SNF4 | | Authors: | Rudolph, M.J, Amodeo, G.A, Iram, S, Hong, S, Pirino, G, Carlson, M, Tong, L. | | Deposit date: | 2006-11-20 | | Release date: | 2006-12-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Bateman2 domain of yeast Snf4: dimeric association and relevance for AMP binding.
Structure, 15, 2007
|
|
2NYC
 
 | | Crystal structure of the Bateman2 domain of yeast Snf4 | | Descriptor: | Nuclear protein SNF4 | | Authors: | Rudolph, M.J, Amodeo, G.A, Iram, S, Hong, S, Pirino, G, Carlson, M, Tong, L. | | Deposit date: | 2006-11-20 | | Release date: | 2006-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Bateman2 domain of yeast Snf4: dimeric association and relevance for AMP binding.
Structure, 15, 2007
|
|
2P9M
 
 | | Crystal structure of conserved hypothetical protein MJ0922 from Methanocaldococcus jannaschii DSM 2661 | | Descriptor: | Hypothetical protein MJ0922 | | Authors: | Zhao, M, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Zhu, J, Swindell II, J.T, Chen, L, Fu, Z.-Q, Charz, J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-26 | | Release date: | 2007-07-03 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of conserved hypothetical protein MJ0922 from Methanocaldococcus jannaschii DSM 2661
To be Published
|
|
2PFI
 
 | |
