1UI0
 
 | | Crystal Structure Of Uracil-DNA Glycosylase From Thermus Thermophilus HB8 | | Descriptor: | IRON/SULFUR CLUSTER, SULFATE ION, URACIL, ... | | Authors: | Hoseki, J, Okamoto, A, Masui, R, Shibata, T, Inoue, Y, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-14 | | Release date: | 2003-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of a Family 4 Uracil-DNA Glycosylase from Thermus thermophilus HB8
J.Mol.Biol., 333, 2003
|
|
1UI1
 
 | | Crystal Structure Of Uracil-DNA Glycosylase From Thermus Thermophilus HB8 | | Descriptor: | IRON/SULFUR CLUSTER, Uracil-DNA Glycosylase | | Authors: | Hoseki, J, Okamoto, A, Masui, R, Shibata, T, Inoue, Y, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-14 | | Release date: | 2003-10-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of a Family 4 Uracil-DNA Glycosylase from Thermus thermophilus HB8
J.Mol.Biol., 333, 2003
|
|
1UUG
 
 | | ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE:INHIBITOR COMPLEX WITH WILD-TYPE UDG AND WILD-TYPE UGI | | Descriptor: | URACIL-DNA GLYCOSYLASE, URACIL-DNA GLYCOSYLASE INHIBITOR | | Authors: | Mol, C.D, Arvai, A.S, Putnam, C.D, Tainer, J.A. | | Deposit date: | 1998-10-31 | | Release date: | 1999-03-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase
J.Mol.Biol., 287, 1999
|
|
1VK2
 
 | |
1WYW
 
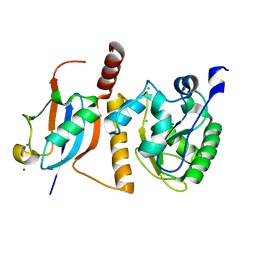 | | Crystal Structure of SUMO1-conjugated thymine DNA glycosylase | | Descriptor: | CHLORIDE ION, G/T mismatch-specific thymine DNA glycosylase, MAGNESIUM ION, ... | | Authors: | Baba, D, Maita, N, Jee, J.G, Uchimura, Y, Saitoh, H, Sugasawa, K, Hanaoka, F, Tochio, H, Hiroaki, H, Shirakawa, M. | | Deposit date: | 2005-02-17 | | Release date: | 2005-06-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of thymine DNA glycosylase conjugated to SUMO-1.
Nature, 435, 2005
|
|
1YUO
 
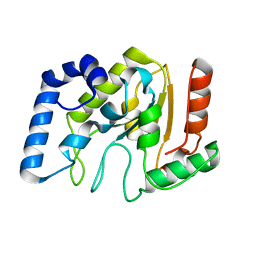 | | Optimisation of the surface electrostatics as a strategy for cold adaptation of uracil-DNA N-glycosylase (UNG)from atlantic cod (Gadus morhua) | | Descriptor: | Uracil-DNA glycosylase | | Authors: | Moe, E, Leiros, I, Riise, E.K, Olufsen, M, Lanes, O, Smalas, A.O, Willassen, N.P. | | Deposit date: | 2005-02-14 | | Release date: | 2005-03-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Optimisation of the surface electrostatics as a strategy for cold adaptation of uracil-DNA N-glycosylase (UNG) from Atlantic cod (Gadus morhua)
J.Mol.Biol., 343, 2004
|
|
2BOO
 
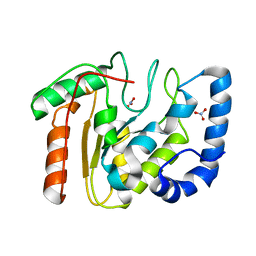 | | The crystal structure of Uracil-DNA N-Glycosylase (UNG) from Deinococcus radiodurans. | | Descriptor: | NITRATE ION, URACIL-DNA GLYCOSYLASE | | Authors: | Leiros, I, Moe, E, Smalas, A.O, McSweeney, S. | | Deposit date: | 2005-04-13 | | Release date: | 2005-07-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Uracil-DNA N-Glycosylase (Ung) from Deinococcus Radiodurans.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2C2P
 
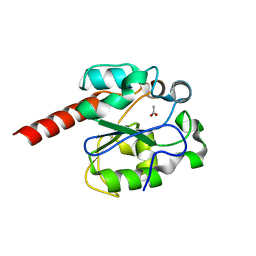 | | The crystal structure of mismatch specific uracil-DNA glycosylase (MUG) from Deinococcus radiodurans | | Descriptor: | ACETATE ION, G/U MISMATCH-SPECIFIC DNA GLYCOSYLASE | | Authors: | Moe, E, Leiros, I, Smalas, A.O, McSweeney, S. | | Deposit date: | 2005-09-29 | | Release date: | 2005-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Crystal Structure of Mismatch Specific Uracil-DNA Glycosylase (Mug) from Deinococcus Radiodurans Reveals a Novel Catalytic Residue and Broad Substrate Specificity
J.Biol.Chem., 281, 2006
|
|
2C2Q
 
 | | The crystal structure of mismatch specific uracil-DNA glycosylase (MUG) from Deinococcus radiodurans. Inactive mutant Asp93Ala. | | Descriptor: | ACETATE ION, G/U MISMATCH-SPECIFIC DNA GLYCOSYLASE | | Authors: | Moe, E, Leiros, I, Smalas, A.O, McSweeney, S. | | Deposit date: | 2005-09-29 | | Release date: | 2005-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of Mismatch Specific Uracil-DNA Glycosylase (Mug) from Deinococcus Radiodurans Reveals a Novel Catalytic Residue and Broad Substrate Specificity
J.Biol.Chem., 281, 2006
|
|
2C53
 
 | | A comparative study of uracil DNA glycosylases from human and herpes simplex virus type 1 | | Descriptor: | 2'-DEOXYURIDINE, GLYCEROL, URACIL DNA GLYCOSYLASE | | Authors: | Krusong, K, Carpenter, E.P, Bellmy, S.R.W, Savva, R, Baldwin, G.S. | | Deposit date: | 2005-10-25 | | Release date: | 2005-11-28 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Comparative Study of Uracil-DNA Glycosylases from Human and Herpes Simplex Virus Type 1.
J.Biol.Chem., 281, 2006
|
|
2C56
 
 | | A comparative study of uracil DNA glycosylases from human and herpes simplex virus type 1 | | Descriptor: | URACIL DNA GLYCOSYLASE, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Krusong, K, Carpenter, E.P, Bellamy, S.R.W, Savva, R, Baldwin, G.S. | | Deposit date: | 2005-10-26 | | Release date: | 2005-11-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Comparative Study of Uracil-DNA Glycosylases from Human and Herpes Simplex Virus Type 1.
J.Biol.Chem., 281, 2006
|
|
2D07
 
 | | Crystal Structure of SUMO-3-modified Thymine-DNA Glycosylase | | Descriptor: | G/T mismatch-specific thymine DNA glycosylase, Ubiquitin-like protein SMT3B | | Authors: | Baba, D, Maita, N, Jee, J.G, Uchimura, Y, Saitoh, H, Sugasawa, K, Hanaoka, F, Tochio, H, Hiroaki, H, Shirakawa, M. | | Deposit date: | 2005-07-26 | | Release date: | 2006-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of SUMO-3-modified Thymine-DNA Glycosylase
J.Mol.Biol., 359, 2006
|
|
2D3Y
 
 | | Crystal structure of uracil-DNA glycosylase from Thermus Thermophilus HB8 | | Descriptor: | 2'-DEOXYURIDINE-5'-MONOPHOSPHATE, ACETATE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Kosaka, H, Nakagawa, N, Masui, R, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-10-04 | | Release date: | 2006-10-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of family 5 uracil-DNA glycosylase bound to DNA.
J.Mol.Biol., 373, 2007
|
|
2DDG
 
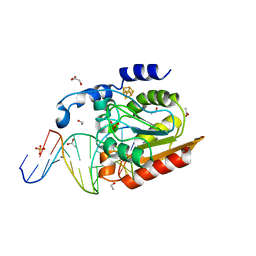 | | Crystal structure of uracil-DNA glycosylase in complex with AP:G containing DNA | | Descriptor: | 5'-D(*AP*TP*GP*TP*TP*GP*CP*(D1P)P*TP*TP*AP*GP*TP*CP*C)-3', 5'-D(*GP*GP*AP*CP*TP*AP*AP*GP*GP*CP*AP*AP*CP*A)-3', ACETATE ION, ... | | Authors: | Kosaka, H, Nakagawa, N, Masui, R, Hoseki, J, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-01-28 | | Release date: | 2007-02-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of family 5 uracil-DNA glycosylase bound to DNA.
J.Mol.Biol., 373, 2007
|
|
2DEM
 
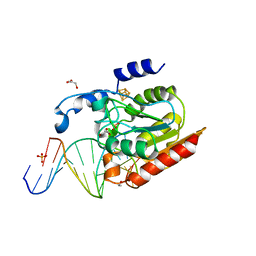 | | Crystal structure of Uracil-DNA glycosylase in complex with AP:A containing DNA | | Descriptor: | 5'-D(*AP*TP*GP*TP*TP*GP*CP*(D1P)P*TP*TP*AP*GP*TP*CP*C)-3', 5'-D(*GP*GP*AP*CP*TP*AP*AP*AP*GP*CP*AP*AP*CP*A)-3', DIHYDROGENPHOSPHATE ION, ... | | Authors: | Kosaka, H, Nakagawa, N, Masui, R, Hoseki, J, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-02-13 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of family 5 uracil-DNA glycosylase bound to DNA.
J.Mol.Biol., 373, 2007
|
|
2DP6
 
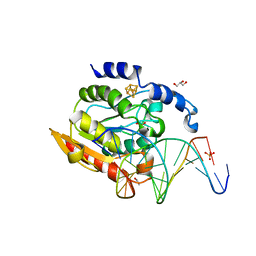 | | Crystal structure of uracil-DNA glycosylase in complex with AP:C containing DNA | | Descriptor: | 5'-D(*AP*TP*GP*TP*TP*GP*CP*(D1P)P*TP*TP*AP*GP*TP*CP*C)-3', 5'-D(*GP*GP*AP*CP*TP*AP*AP*CP*GP*CP*AP*AP*CP*A)-3', DIHYDROGENPHOSPHATE ION, ... | | Authors: | Kosaka, H, Nakagawa, N, Masui, R, Kuramitsu, S, Hoseki, J, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-05-07 | | Release date: | 2007-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Family 5 Uracil-DNA Glycosylase Bound to DNA Reveals Insights into the Mechanism for Substrate Recognition and Catalysis
To be Published
|
|
2EUG
 
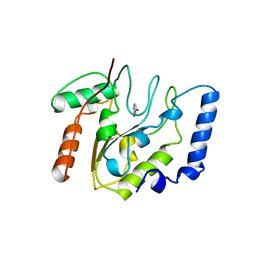 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI URACIL DNA GLYCOSYLASE AND ITS COMPLEXES WITH URACIL AND GLYCEROL: STRUCTURE AND GLYCOSYLASE MECHANISM REVISITED | | Descriptor: | PROTEIN (GLYCOSYLASE), URACIL | | Authors: | Xiao, G, Tordova, M, Jagadeesh, J, Drohat, A.C, Stivers, J.T, Gilliland, G.L. | | Deposit date: | 1998-10-13 | | Release date: | 1999-10-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of Escherichia coli uracil DNA glycosylase and its complexes with uracil and glycerol: structure and glycosylase mechanism revisited.
Proteins, 35, 1999
|
|
2HXM
 
 | | Complex of UNG2 and a small Molecule synthetic Inhibitor | | Descriptor: | 4-[(1E,7E)-8-(2,6-DIOXO-1,2,3,6-TETRAHYDROPYRIMIDIN-4-YL)-3,6-DIOXA-2,7-DIAZAOCTA-1,7-DIEN-1-YL]BENZOIC ACID, Uracil-DNA glycosylase | | Authors: | Bianchet, M.A, Krosky, D.J, Ghung, S, Seiple, L, Amzel, L.M, Stivers, J.T. | | Deposit date: | 2006-08-03 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mimicking damaged DNA with a small molecule inhibitor of human UNG2.
Nucleic Acids Res., 34, 2006
|
|
2J8X
 
 | | Epstein-Barr virus uracil-DNA glycosylase in complex with Ugi from PBS-2 | | Descriptor: | URACIL-DNA GLYCOSYLASE, URACIL-DNA GLYCOSYLASE INHIBITOR, UREA | | Authors: | Geoui, T, Buisson, M, Tarbouriech, N, Burmeister, W.P. | | Deposit date: | 2006-10-31 | | Release date: | 2006-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | New Insights on the Role of the Gamma-Herpesvirus Uracil-DNA Glycosylase Leucine Loop Revealed by the Structure of the Epstein-Barr Virus Enzyme in Complex with an Inhibitor Protein.
J.Mol.Biol., 366, 2007
|
|
2JHQ
 
 | | Crystal structure of Uracil DNA-glycosylase from Vibrio cholerae | | Descriptor: | CHLORIDE ION, URACIL DNA-GLYCOSYLASE | | Authors: | Raeder, I.L.U, Moe, E, Willassen, N.P, Smalas, A.O, Leiros, I. | | Deposit date: | 2007-02-23 | | Release date: | 2008-06-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Uracil-DNA N-Glycosylase (Ung) from Vibrio Cholerae. Mapping Temperature Adaptation Through Structural and Mutational Analysis.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2L3F
 
 | | Solution NMR Structure of a putative Uracil DNA glycosylase from Methanosarcina acetivorans, Northeast Structural Genomics Consortium Target MvR76 | | Descriptor: | Uncharacterized protein | | Authors: | Aramini, J.M, Hamilton, K, Ciccosanti, C.T, Wang, H, Lee, H.W, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-09-13 | | Release date: | 2010-10-20 | | Last modified: | 2012-02-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of a putative Uracil DNA glycosylase from Methanosarcina acetivorans, Northeast Structural Genomics Consortium Target MvR76
To be Published
|
|
2OXM
 
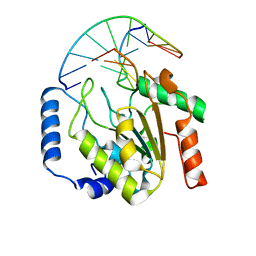 | | Crystal structure of a UNG2/modified DNA complex that represent a stabilized short-lived extrahelical state in ezymatic DNA base flipping | | Descriptor: | DNA (5'-D(*AP*AP*AP*GP*AP*TP*(4MF)P*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*TP*AP*TP*CP*TP*T)-3'), Uracil-DNA glycosylase | | Authors: | Bianchet, M.A, Krosky, D.J, Stivers, J.T, Amzel, L.M. | | Deposit date: | 2007-02-20 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Enzymatic capture of an extrahelical thymine in the search for uracil in DNA.
Nature, 449, 2007
|
|
2OYT
 
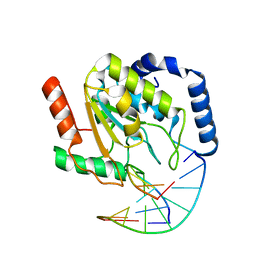 | | Crystal Structure of UNG2/DNA(TM) | | Descriptor: | DNA strand1, DNA strand2, Uracil-DNA glycosylase | | Authors: | Bianchet, M.A, Krosky, D.J, Stivers, J.T, Amzel, L.M. | | Deposit date: | 2007-02-22 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enzymatic capture of an extrahelical thymine in the search for uracil in DNA.
Nature, 449, 2007
|
|
2RBA
 
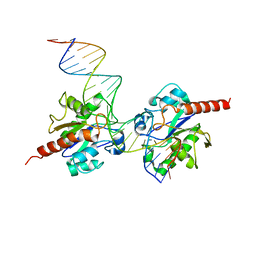 | | Structure of Human Thymine DNA Glycosylase Bound to Abasic and Undamaged DNA | | Descriptor: | DNA (5'-D(*DCP*DAP*DGP*DCP*DTP*DCP*DTP*DGP*DTP*DAP*DCP*DGP*DTP*DGP*DAP*DGP*DCP*DAP*DGP*DTP*DGP*DGP*DA)-3'), DNA (5'-D(*DCP*DCP*DAP*DCP*DTP*DGP*DCP*DTP*DCP*DAP*(3DR)P*DGP*DTP*DAP*DCP*DAP*DGP*DAP*DGP*DCP*DTP*DGP*DT)-3'), G/T mismatch-specific thymine DNA glycosylase | | Authors: | Maiti, A, Pozharski, E, Drohat, A.C. | | Deposit date: | 2007-09-18 | | Release date: | 2008-06-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal structure of human thymine DNA glycosylase bound to DNA elucidates sequence-specific mismatch recognition.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2SSP
 
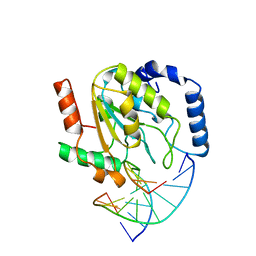 | | LEUCINE-272-ALANINE URACIL-DNA GLYCOSYLASE BOUND TO ABASIC SITE-CONTAINING DNA | | Descriptor: | DNA (5'-D(*AP*AP*AP*GP*AP*TP*AP*AP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*TP*(AAB)P*AP*TP*CP*TP*T)-3'), PROTEIN (URACIL-DNA GLYCOSYLASE) | | Authors: | Parikh, S.S, Mol, C.D, Slupphaug, G, Bharati, S, Krokan, H.E, Tainer, J.A. | | Deposit date: | 1999-04-28 | | Release date: | 1999-05-06 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA.
EMBO J., 17, 1998
|
|
