8YI6
 
 | | BesA wild-type from Streptomyces cattleyicolor | | Descriptor: | ADENINE, L-propargylglycine--L-glutamate ligase | | Authors: | Fujishiro, T. | | Deposit date: | 2024-02-29 | | Release date: | 2025-03-05 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure-guided understanding enzymatic mechanism and substrate specificity of ATP-dependent alkyne-containing dipeptide synthetase BesA
To be published
|
|
8YRM
 
 | | Iota toxin Ib pore serine-clamp mutant | | Descriptor: | CALCIUM ION, Iota toxin component Ib | | Authors: | Ninomiya, Y, Yoshida, T, Yamada, T, Kishikawa, J, Tsuge, H. | | Deposit date: | 2024-03-21 | | Release date: | 2025-03-26 | | Last modified: | 2025-08-06 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | Serine clamp of Clostridium perfringens binary toxin BECb (CPILEb)-pore confers cytotoxicity and enterotoxicity.
Commun Biol, 8, 2025
|
|
8ZCA
 
 | | Crystal structure of human CD47 ECD bound to Fab of Hu1C8 | | Descriptor: | 1C8 Fab heavy chain, 1C8 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, Z, Ding, J. | | Deposit date: | 2024-04-29 | | Release date: | 2025-05-07 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An anti-CD47 antibody binds to a distinct epitope in a novel metal ion-dependent manner to minimize cross-linking of red blood cells.
J.Biol.Chem., 301, 2025
|
|
8ZLB
 
 | | EB bound state of hPAC | | Descriptor: | 4-azanyl-6-[[4-[4-[(~{E})-(8-azanyl-1-oxidanyl-5,7-disulfo-naphthalen-2-yl)diazenyl]-3-methyl-phenyl]-2-methyl-phenyl]diazenyl]-5-oxidanyl-naphthalene-1,3-disulfonic acid, Proton-activated chloride channel | | Authors: | Su, N, Chen, X. | | Deposit date: | 2024-05-18 | | Release date: | 2025-07-23 | | Last modified: | 2025-08-06 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Discovery and structural basis of endogenous and exogenous inhibitors of the proton-activated-chloride channel.
Cell Rep, 44, 2025
|
|
8ZLL
 
 | | The apo state of hPAC with endogenous cholesterol | | Descriptor: | CHOLESTEROL, Proton-activated chloride channel | | Authors: | Zhang, H, Chen, X. | | Deposit date: | 2024-05-20 | | Release date: | 2025-07-23 | | Last modified: | 2025-08-06 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Discovery and structural basis of endogenous and exogenous inhibitors of the proton-activated-chloride channel.
Cell Rep, 44, 2025
|
|
8ZUJ
 
 | | Pentagonal cluster of BAFF-BAFFR ectodomain complex | | Descriptor: | Tumor necrosis factor ligand superfamily member 13b, soluble form, Tumor necrosis factor receptor superfamily member 13C | | Authors: | Lim, C.S, Lee, J.O. | | Deposit date: | 2024-06-09 | | Release date: | 2025-06-11 | | Last modified: | 2025-08-06 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Highly ordered clustering of TNF alpha and BAFF ligand-receptor-intracellular adaptor complexes on a lipid membrane.
Nat Commun, 16, 2025
|
|
8ZUK
 
 | | Cluster structure of the BAFF-BAFFR-TRAF3 complex | | Descriptor: | TNF receptor-associated factor 3, Tumor necrosis factor receptor superfamily member 13C | | Authors: | Lim, C.S, Lee, J.O. | | Deposit date: | 2024-06-09 | | Release date: | 2025-06-11 | | Last modified: | 2025-08-06 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Highly ordered clustering of TNF alpha and BAFF ligand-receptor-intracellular adaptor complexes on a lipid membrane.
Nat Commun, 16, 2025
|
|
9ATV
 
 | | X-ray co-crystal structure of NCGC00685960 / NCATS-SM9335 in NNMT | | Descriptor: | (8M)-8-{3-[(3S)-1-(2-cyclohexylethyl)piperidin-3-yl]-5-oxo-4,5-dihydro-1H-1,2,4-triazol-1-yl}quinolin-2(1H)-one, Nicotinamide N-methyltransferase | | Authors: | Olland, A, White, A, Suto, R. | | Deposit date: | 2024-02-27 | | Release date: | 2025-03-05 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (2.413 Å) | | Cite: | NNMT inhibition in cancer-associated fibroblasts restores antitumour immunity.
Nature, 2025
|
|
9B2S
 
 | | Haspin bound to nucleosome in position 1 | | Descriptor: | 601 DNA (185-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Hicks, C.W, Wolberger, C. | | Deposit date: | 2024-03-16 | | Release date: | 2025-01-22 | | Last modified: | 2025-08-06 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Haspin kinase binds to a nucleosomal DNA supergroove.
Nat.Struct.Mol.Biol., 32, 2025
|
|
9B4U
 
 | | Crystal structure of p110alpha-RBD covalently bound to a breaker compound BBO-10203 via Cys242 | | Descriptor: | 1-[(4R,8R)-2-[(4M,7P)-7-[2,4-difluoro-6-(2-methoxyethoxy)phenyl]-4-(1-methyl-1H-indazol-5-yl)thieno[3,2-c]pyridin-6-yl]-4-methyl-6,7-dihydropyrazolo[1,5-a]pyrazin-5(4H)-yl]propan-1-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Czyzyk, D.J, Simanshu, D.K. | | Deposit date: | 2024-03-21 | | Release date: | 2025-06-25 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | BBO-10203 inhibits tumor growth without inducing hyperglycemia by blocking RAS-PI3K alpha interaction.
Science, 389, 2025
|
|
9BW5
 
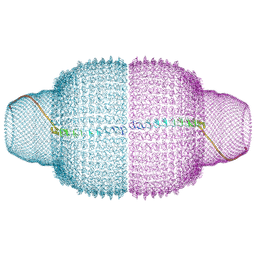 | | Human Vault Cage | | Descriptor: | Major vault protein | | Authors: | Lodwick, J.E, Zhao, M. | | Deposit date: | 2024-05-20 | | Release date: | 2025-02-05 | | Last modified: | 2025-08-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into the roles of PARP4 and NAD + binding in the human vault cage.
Nat Commun, 16, 2025
|
|
9BW6
 
 | | Human Vault Cage in complex with PARP4 | | Descriptor: | Major vault protein, Protein mono-ADP-ribosyltransferase PARP4 | | Authors: | Lodwick, J.E, Zhao, M. | | Deposit date: | 2024-05-20 | | Release date: | 2025-02-05 | | Last modified: | 2025-08-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into the roles of PARP4 and NAD + binding in the human vault cage.
Nat Commun, 16, 2025
|
|
9BW7
 
 | | Human Vault Cage in complex with PARP4 and NAD+ | | Descriptor: | Major vault protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Protein mono-ADP-ribosyltransferase PARP4 | | Authors: | Lodwick, J.E, Zhao, M. | | Deposit date: | 2024-05-20 | | Release date: | 2025-02-05 | | Last modified: | 2025-08-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into the roles of PARP4 and NAD + binding in the human vault cage.
Nat Commun, 16, 2025
|
|
9BWA
 
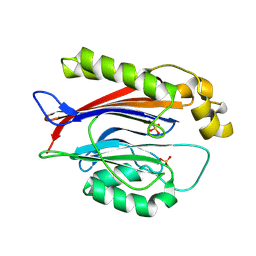 | | Crystal structure of the Transport and Golgi Organization protein 2 Homolog (TANGO2) | | Descriptor: | PHOSPHATE ION, Transport and Golgi organization protein 2 homolog | | Authors: | Lovell, S, Cooper, A, Powers, A, Battaile, K.P, Mohsen, A.-W, Ghaloul-Gonzalez, L. | | Deposit date: | 2024-05-21 | | Release date: | 2025-04-30 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of Human Transport and Golgi Organization 2 Homolog (TANGO2) Protein Reveals an alpha beta beta alpha-Fold Arrangement.
Proteins, 2025
|
|
9C0P
 
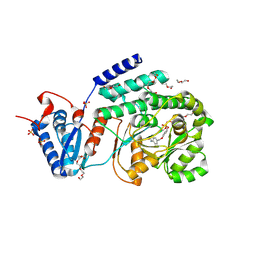 | | M. tuberculosis PKS13 acyltransferase (AT) domain in complex with SuFEx inhibitor CEC215 | | Descriptor: | N-{4-[(4-{[fluorodi(hydroxy)-lambda~4~-sulfanyl]oxy}phenoxy)methyl]phenyl}acetamide, PENTAETHYLENE GLYCOL, Polyketide synthase Pks13, ... | | Authors: | Krieger, I.K, Tang, S, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2024-05-27 | | Release date: | 2025-05-07 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | SuFEx-based antitubercular compound irreversibly inhibits Pks13
Nature, 2025
|
|
9C1C
 
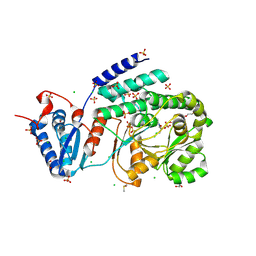 | |
9C1D
 
 | | Mycobaterium tuberculosis Pks13 acyltransferase incubated with DMSO | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ... | | Authors: | Tang, S, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2024-05-29 | | Release date: | 2025-05-21 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | SuFEx-based antitubercular compound irreversibly inhibits Pks13
Nature, 2025
|
|
9C1V
 
 | | M. tuberculosis PKS13 acyltransferase (AT) domain in complex with SuFEx inhibitor CMX410 | | Descriptor: | N-(1-{3,5-difluoro-4-[(4-{[fluorodi(hydroxy)-lambda~4~-sulfanyl]oxy}phenoxy)methyl]phenyl}-1H-1,2,4-triazol-3-yl)methanesulfonamide, PENTAETHYLENE GLYCOL, Polyketide synthase Pks13, ... | | Authors: | Krieger, I.K, Tang, S, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2024-05-29 | | Release date: | 2025-05-07 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | SuFEx-based antitubercular compound irreversibly inhibits Pks13
Nature, 2025
|
|
9C1Y
 
 | |
9C1Z
 
 | |
9C2R
 
 | | M. tuberculosis PKS13 acyltransferase (AT) domain sulfate free apo form | | Descriptor: | PENTAETHYLENE GLYCOL, Polyketide synthase Pks13, SULFATE ION, ... | | Authors: | Krieger, I.K, Tang, S, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2024-05-31 | | Release date: | 2025-05-07 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | SuFEx-based antitubercular compound irreversibly inhibits Pks13
Nature, 2025
|
|
9C5W
 
 | |
9C9O
 
 | | M. tuberculosis PKS13 acyltransferase (AT) domain in complex with SuFEx inhibitor CMX410 - reaction product | | Descriptor: | 4-({2,6-difluoro-4-[3-(methanesulfonamido)-1H-1,2,4-triazol-1-yl]phenyl}methoxy)phenyl hydrogen sulfate, PENTAETHYLENE GLYCOL, Polyketide synthase Pks13, ... | | Authors: | Krieger, I.V, Tang, S, Sacchetini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2024-06-14 | | Release date: | 2025-05-07 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | SuFEx-based antitubercular compound irreversibly inhibits Pks13
Nature, 2025
|
|
9CMK
 
 | | Crystal structure of p110alpha-RAS binding domain (RBD) in complex with molecular glue D927 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[3-fluoro-4-({(7P)-7-[2-(2-methoxyethoxy)phenyl]thieno[2,3-d]pyridazin-4-yl}amino)phenyl]acetamide, ... | | Authors: | Czyzyk, D.J, Simanshu, D.K. | | Deposit date: | 2024-07-15 | | Release date: | 2025-07-23 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular glues that facilitate RAS binding to PI3K alpha promote glucose uptake without insulin.
Science, 389, 2025
|
|
9CML
 
 | |
