9DEZ
 
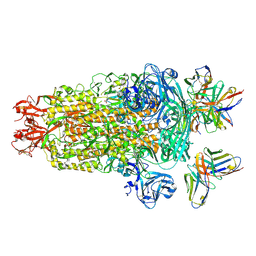 | | PDCoV S trimer bound by three copies of PD41 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITOLEIC ACID, ... | | Authors: | Asarnow, D, Rexhepaj, M, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2024-08-29 | | Release date: | 2024-11-13 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Isolation and escape mapping of broadly neutralizing antibodies against emerging delta-coronaviruses.
Immunity, 57, 2024
|
|
9DF0
 
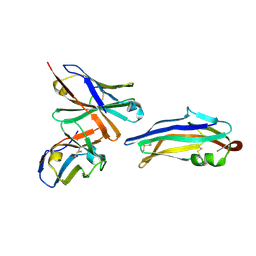 | | PDCoV S RBD bound to PD41 Fab (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PD41 Fab variable heavy-chain, PD41 Fab variable light-chain, ... | | Authors: | Asarnow, D, Rexhepaj, M, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2024-08-29 | | Release date: | 2024-11-13 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Isolation and escape mapping of broadly neutralizing antibodies against emerging delta-coronaviruses.
Immunity, 57, 2024
|
|
9DWY
 
 | | Cryo-EM structure of human MCT8 in complex with thyroid hormone T3 | | Descriptor: | 3,5,3'TRIIODOTHYRONINE, Monocarboxylate transporter 8 | | Authors: | Ge, Y, Dou, T, Jiang, J, Huang, P. | | Deposit date: | 2024-10-10 | | Release date: | 2025-07-16 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into brain thyroid hormone transport via MCT8 and OATP1C1.
Cell, 2025
|
|
9DXO
 
 | |
9DXP
 
 | | Cryo-EM structure of human OATP1C1 in complex with thyroid hormone T4 | | Descriptor: | 3,5,3',5'-TETRAIODO-L-THYRONINE, CHOLESTEROL HEMISUCCINATE, Solute carrier organic anion transporter family member 1C1 | | Authors: | Ge, Y, Dou, T, Jiang, J, Huang, P. | | Deposit date: | 2024-10-11 | | Release date: | 2025-07-16 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural insights into brain thyroid hormone transport via MCT8 and OATP1C1.
Cell, 2025
|
|
9DXX
 
 | |
9E6Q
 
 | |
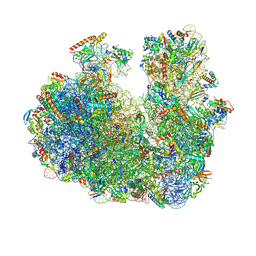
9E71
 
 | |
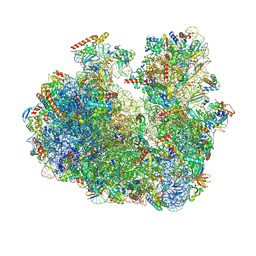
9E7F
 
 | |
9ELY
 
 | | raiA RNA motif (Clostridium acetobutylicum) | | Descriptor: | raiA RNA | | Authors: | Kretsch, R.C, Wu, Y, Nye, G, Das, R, Chiu, W. | | Deposit date: | 2024-12-05 | | Release date: | 2025-04-23 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Naturally ornate RNA-only complexes revealed by cryo-EM.
Nature, 643, 2025
|
|
9F5W
 
 | | Human condensin II - M18BP1 complex | | Descriptor: | Condensin-2 complex subunit D3, Condensin-2 complex subunit G2, Condensin-2 complex subunit H2, ... | | Authors: | Borsellini, A, Vannini, A. | | Deposit date: | 2024-04-30 | | Release date: | 2025-07-02 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Condensin II activation by M18BP1.
Mol.Cell, 85, 2025
|
|
9GFK
 
 | | human MDM2 complex with stapled foldamer | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, SULFATE ION, Stapled foldamer | | Authors: | Neuville, M, Bourgeais, M, Buratto, J, Saragaglia, C, Bo, L, Mauran, L, Varajao, L, Goudreau, S.R, Kauffmann, B, Thinon, E, Pasco, M, Khatib, A.M. | | Deposit date: | 2024-08-09 | | Release date: | 2025-03-26 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (1.841 Å) | | Cite: | Optimal Stapling of a Helical Peptide-Foldamer Hybrid Using a C-Terminal 4-Mercaptoproline Enhances Protein Surface Recognition and Cellular Activity.
Chemistry, 31, 2025
|
|
9GTG
 
 | | RIPK1 in complex with AZ"902 | | Descriptor: | (5R)-5-[(7-chloro-1H-indol-3-yl)methyl]-3-methylimidazolidine-2,4-dione, Receptor-interacting serine/threonine-protein kinase 1, ~{N}-[[(3~{S})-1-ethanoylpyrrolidin-3-yl]methyl]-~{N}-methyl-4-quinolin-7-yl-benzenesulfonamide | | Authors: | Petersen, J. | | Deposit date: | 2024-09-17 | | Release date: | 2025-07-02 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Discovery and Validation of a Novel Class of Necroptosis Inhibitors Targeting RIPK1.
Acs Chem.Biol., 20, 2025
|
|
9GTY
 
 | | RIPK1 in complex with AZ"320 | | Descriptor: | (5R)-5-[(7-chloro-1H-indol-3-yl)methyl]-3-methylimidazolidine-2,4-dione, 7-[4-[3-(methylsulfonylmethyl)azetidin-1-yl]sulfonylphenyl]quinoline, DIMETHYL SULFOXIDE, ... | | Authors: | Petersen, J. | | Deposit date: | 2024-09-18 | | Release date: | 2025-07-16 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (2.145 Å) | | Cite: | Discovery and Validation of a Novel Class of Necroptosis Inhibitors Targeting RIPK1.
Acs Chem.Biol., 20, 2025
|
|
9GYD
 
 | | Ferredoxin Wild-type -As-isolated state | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Ferredoxin-1, chloroplastic | | Authors: | Carr, S.B, Wei, J, Vincent, K.A. | | Deposit date: | 2024-10-01 | | Release date: | 2025-05-28 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Cyanophenylalanine as an Infrared Probe for Iron-Sulfur Cluster Redox State in Multicenter Metalloenzymes.
Chembiochem, 26, 2025
|
|
9GYL
 
 | | Ferredoxin Wild-type -Oxidised state | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Ferredoxin-1, chloroplastic, ... | | Authors: | Carr, S.B, Wei, J, Vincent, K.A. | | Deposit date: | 2024-10-02 | | Release date: | 2025-05-28 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Cyanophenylalanine as an Infrared Probe for Iron-Sulfur Cluster Redox State in Multicenter Metalloenzymes.
Chembiochem, 26, 2025
|
|
9GYN
 
 | | Ferredoxin Wild-type - Reduced state | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Ferredoxin-1, chloroplastic | | Authors: | Carr, S.B, Wei, J, Vincent, K.A. | | Deposit date: | 2024-10-02 | | Release date: | 2025-05-28 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Cyanophenylalanine as an Infrared Probe for Iron-Sulfur Cluster Redox State in Multicenter Metalloenzymes.
Chembiochem, 26, 2025
|
|
9GYR
 
 | | Ferredoxin CNF labelled, oxidised state | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Ferredoxin-1, chloroplastic | | Authors: | Carr, S.B, Wei, J, Vincent, K.A. | | Deposit date: | 2024-10-02 | | Release date: | 2025-05-28 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Cyanophenylalanine as an Infrared Probe for Iron-Sulfur Cluster Redox State in Multicenter Metalloenzymes.
Chembiochem, 26, 2025
|
|
9GYU
 
 | | Ferredoxin CNF-labelled reduced state | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Ferredoxin-1, chloroplastic | | Authors: | Carr, S.B, Wei, J, Vincent, K.A. | | Deposit date: | 2024-10-03 | | Release date: | 2025-05-28 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Cyanophenylalanine as an Infrared Probe for Iron-Sulfur Cluster Redox State in Multicenter Metalloenzymes.
Chembiochem, 26, 2025
|
|
9GZ0
 
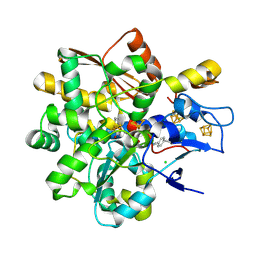 | | FeFe Hydrogenase from Desulfovibrio desulfuricans labelled with cyanophenylalanine - oxidised state | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, LITHIUM ION, ... | | Authors: | Carr, S.B, Duan, Z, Rodroguez-Macia, P, Vincent, K.A. | | Deposit date: | 2024-10-03 | | Release date: | 2025-05-28 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Cyanophenylalanine as an Infrared Probe for Iron-Sulfur Cluster Redox State in Multicenter Metalloenzymes.
Chembiochem, 26, 2025
|
|
9GZ4
 
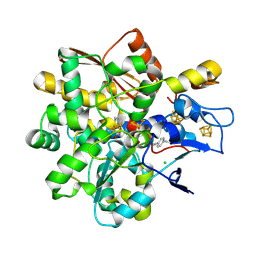 | | FeFe Hydrogenase from Desulfovibrio desulfuricans labelled with cyanophenylalanine - reduced state | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, LITHIUM ION, ... | | Authors: | Carr, S.B, Duan, Z, Rodroguez-Macia, P, Vincent, K.A. | | Deposit date: | 2024-10-03 | | Release date: | 2025-05-28 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Cyanophenylalanine as an Infrared Probe for Iron-Sulfur Cluster Redox State in Multicenter Metalloenzymes.
Chembiochem, 26, 2025
|
|
9GZL
 
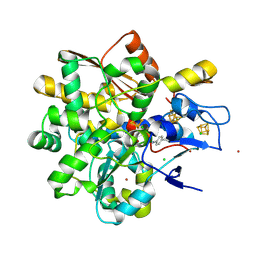 | | Apo FeFe Hydrogenase from Desulfovibrio desulfuricans labelled with cyanophenylalanine | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, LITHIUM ION, ... | | Authors: | Carr, S.B, Duan, Z, Rodriguez-Macia, P, Vincent, K.A. | | Deposit date: | 2024-10-04 | | Release date: | 2025-05-28 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Cyanophenylalanine as an Infrared Probe for Iron-Sulfur Cluster Redox State in Multicenter Metalloenzymes.
Chembiochem, 26, 2025
|
|
9HEJ
 
 | | Dimeric CD163 bound to human haptoglobin-haemoglobin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Zhou, R.X, Higgins, M.K. | | Deposit date: | 2024-11-14 | | Release date: | 2025-07-02 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Scavenger receptor CD163 multimerises to allow uptake of diverse ligands.
Nat Commun, 16, 2025
|
|
9HEK
 
 | | Trimeric CD163 bound to human haptoglobin-haemoglobin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Zhou, R.X, Higgins, M.K. | | Deposit date: | 2024-11-14 | | Release date: | 2025-07-02 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Scavenger receptor CD163 multimerises to allow uptake of diverse ligands.
Nat Commun, 16, 2025
|
|
9HEL
 
 | | Unliganded dimeric CD163 with arm-arm contacts | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Zhou, R.X, Higgins, M.K. | | Deposit date: | 2024-11-14 | | Release date: | 2025-07-02 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Scavenger receptor CD163 multimerises to allow uptake of diverse ligands.
Nat Commun, 16, 2025
|
|
