1E30
 
 | |
1H4G
 
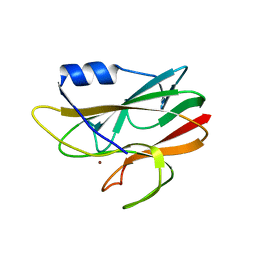
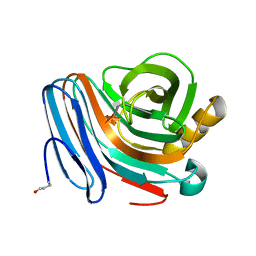 | | Oligosaccharide-binding to family 11 xylanases: both covalent intermediate and mutant-product complexes display 2,5B conformations at the active-centre | | Descriptor: | SULFATE ION, XYLANASE, beta-D-xylopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-xylopyranose | | Authors: | Sabini, E, Wilson, K.S, Danielsen, S, Schulein, M, Davies, G.J. | | Deposit date: | 2001-05-11 | | Release date: | 2002-05-09 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Oligosaccharide binding to family 11 xylanases: both covalent intermediate and mutant product complexes display (2,5)B conformations at the active centre.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1H4H
 
 | | Oligosaccharide-binding to family 11 xylanases: both covalent intermediate and mutant-product complexes display 2,5B conformations at the active-centre | | Descriptor: | XYLANASE, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-xylopyranose | | Authors: | Sabini, E, Wilson, K.S, Danielsen, S, Schulein, M, Davies, G.J. | | Deposit date: | 2001-05-11 | | Release date: | 2002-05-09 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Oligosaccharide binding to family 11 xylanases: both covalent intermediate and mutant product complexes display (2,5)B conformations at the active centre.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1NR8
 
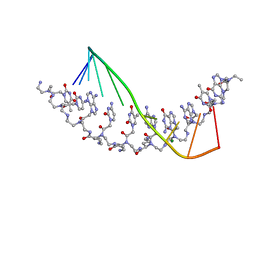 | | The crystal structure of a D-Lysine-based chiral PNA-DNA duplex | | Descriptor: | 5'-D(P*AP*GP*TP*GP*AP*TP*CP*TP*AP*C)-3', H-((GPN)*(TPN)*(APN)*(GPN)*(A66)*(T66)*(C66)*(APN)*(CPN)*(TPN))-NH2, MAGNESIUM ION | | Authors: | Menchise, V, De Simone, G, Tedeschi, T, Corradini, R, Sforza, S, Marchelli, R, Capasso, D, Saviano, M, Pedone, C. | | Deposit date: | 2003-01-24 | | Release date: | 2003-10-28 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Insights into peptide nucleic acid (PNA) structural features: The
crystal structure of a D-lysine-based chiral PNA-DNA duplex
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1P05
 
 | |
1QA0
 
 | | BOVINE TRYPSIN 2-AMINOBENZIMIDAZOLE COMPLEX | | Descriptor: | 1H-benzimidazol-2-amine, CALCIUM ION, TRYPSIN | | Authors: | Whitlow, M. | | Deposit date: | 1999-04-09 | | Release date: | 2000-04-10 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic analysis of potent and selective factor Xa inhibitors complexed to bovine trypsin.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1RMM
 
 | | Probing the Role of Tryptophans in Aequorea Victoria Green Fluorescent Proteins with an Expanded Genetic Code | | Descriptor: | SIGF1-GFP fusion protein | | Authors: | Budisa, N, Pal, P.P, Alefelder, S, Birle, P, Krywcun, T, Rubini, M, Wenger, W, Bae, J.H, Steiner, T. | | Deposit date: | 2003-11-28 | | Release date: | 2004-06-08 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the role of tryptophans in Aequorea victoria green fluorescent proteins with an expanded genetic code
Biol.Chem., 385, 2004
|
|
1RMO
 
 | | Probing the Role of Tryptophans in Aequorea Victoria Green Fluorescent Proteins with an Expanded Genetic Code | | Descriptor: | wunen-nonfunctional GFP fusion protein | | Authors: | Budisa, N, Pal, P.P, Alefelder, S, Birle, P, Krywcun, T, Rubini, M, Wenger, W, Bae, J.H, Steiner, T. | | Deposit date: | 2003-11-28 | | Release date: | 2004-06-08 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Probing the role of tryptophans in Aequorea victoria green fluorescent proteins with an expanded genetic code
Biol.Chem., 385, 2004
|
|
1UB0
 
 | | Crystal Structure Analysis of Phosphomethylpyrimidine Kinase (ThiD) from Thermus Thermophilus Hb8 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, GLYCEROL, Phosphomethylpyrimidine Kinase, ... | | Authors: | Bagautdinov, B, Kuramitsu, S, Yokoyama, S, Miyano, M, Tahirov, T.H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-03-26 | | Release date: | 2003-04-08 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of 4-amino-5-hydroxymethyl-2-methyl pyrimidine phosphate kinase (HMPP kinase) from Thermus thermophilus HB8.
Eur.Biophys.J., 54, 2025
|
|
2FPZ
 
 | | Human tryptase with 2-amino benzimidazole | | Descriptor: | 1H-benzimidazol-2-amine, Tryptase beta-2 | | Authors: | Somoza, J.R. | | Deposit date: | 2006-01-17 | | Release date: | 2006-03-07 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-guided design of Peptide-based tryptase inhibitors.
Biochemistry, 45, 2006
|
|
2FX6
 
 | | bovine trypsin complexed with 2-aminobenzamidazole | | Descriptor: | 1H-benzimidazol-2-amine, CALCIUM ION, Trypsin | | Authors: | Katz, B.A. | | Deposit date: | 2006-02-03 | | Release date: | 2006-02-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structure-guided design of Peptide-based tryptase inhibitors.
Biochemistry, 45, 2006
|
|
3D3D
 
 | | Bacteriophage lambda lysozyme complexed with a chitohexasaccharide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysozyme, SULFATE ION | | Authors: | Leung, A.K.W, Berghuis, A.M. | | Deposit date: | 2008-05-09 | | Release date: | 2008-09-09 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the lytic transglycosylase from bacteriophage lambda in complex with hexa-N-acetylchitohexaose
Biochemistry, 40, 2001
|
|
3H9B
 
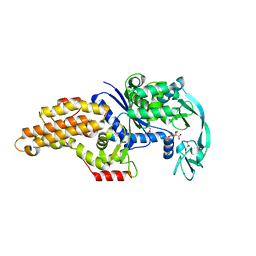 | | Structure of a mutant methionyl-tRNA synthetase with modified specificity complexed with azidonorleucine | | Descriptor: | 6-Azido-L-lysine, CITRIC ACID, Methionyl-tRNA synthetase, ... | | Authors: | Schmitt, E, Tanrikulu, I.C, Yoo, T.H, Panvert, M, Tirrell, D.A, Mechulam, Y. | | Deposit date: | 2009-04-30 | | Release date: | 2009-12-08 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Switching from an induced-fit to a lock-and-key mechanism in an aminoacyl-tRNA synthetase with modified specificity.
J.Mol.Biol., 394, 2009
|
|
3LPR
 
 | |
4MQ9
 
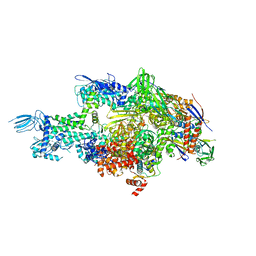 | | Crystal structure of Thermus thermophilus RNA polymerase holoenzyme in complex with GE23077 | | Descriptor: | (2Z)-2-methylbut-2-enoic acid, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Ho, M.X, Arnold, E, Ebright, R.H, Zhang, Y, Tuske, S. | | Deposit date: | 2013-09-16 | | Release date: | 2014-05-07 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
4OIN
 
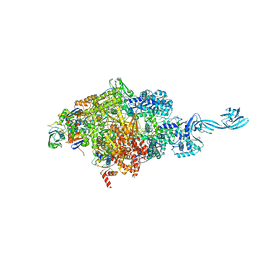 | | Crystal structure of Thermus thermophilus transcription initiation complex soaked with GE23077 | | Descriptor: | (2Z)-2-methylbut-2-enoic acid, 5'-D(*CP*CP*T*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', ... | | Authors: | Zhang, Y, Ebright, R.H, Arnold, E. | | Deposit date: | 2014-01-20 | | Release date: | 2014-05-07 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
4OIP
 
 | | Crystal structure of Thermus thermophilus transcription initiation complex soaked with GE23077, ATP, and CMPcPP | | Descriptor: | (2Z)-2-methylbut-2-enoic acid, 5'-D(*CP*CP*TP*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', ... | | Authors: | Zhang, Y, Ebright, R.H, Arnold, E. | | Deposit date: | 2014-01-20 | | Release date: | 2014-05-07 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
4OIQ
 
 | | Crystal structure of Thermus thermophilus transcription initiation complex soaked with GE23077 and rifampicin | | Descriptor: | (2Z)-2-methylbut-2-enoic acid, 5'-D(*CP*CP*T*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', ... | | Authors: | Zhang, Y, Ebright, R.H, Arnold, E. | | Deposit date: | 2014-01-20 | | Release date: | 2014-05-07 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (3.624 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
4OIR
 
 | | Crystal structure of Thermus thermophilus RNA polymerase transcription initiation complex soaked with GE23077 and rifamycin SV | | Descriptor: | (2Z)-2-methylbut-2-enoic acid, 5'-D(*CP*CP*T*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', ... | | Authors: | Zhang, Y, Ebright, R.H, Arnold, E. | | Deposit date: | 2014-01-20 | | Release date: | 2014-05-07 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (3.105 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
5EC1
 
 | | KcsA with V76ester mutation | | Descriptor: | Antibody Fab Fragment Light Chain, DIACYL GLYCEROL, NONAN-1-OL, ... | | Authors: | Matulef, K, Valiyaveetil, F.I. | | Deposit date: | 2015-10-20 | | Release date: | 2016-04-20 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Individual Ion Binding Sites in the K(+) Channel Play Distinct Roles in C-type Inactivation and in Recovery from Inactivation.
Structure, 24, 2016
|
|
5EC2
 
 | | KcsA with V76ester+G77dA mutations | | Descriptor: | Antibody Fab Fragment Light Chain, DIACYL GLYCEROL, NONAN-1-OL, ... | | Authors: | Matulef, K, Valiyaveetil, F.I. | | Deposit date: | 2015-10-20 | | Release date: | 2016-04-20 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Individual Ion Binding Sites in the K(+) Channel Play Distinct Roles in C-type Inactivation and in Recovery from Inactivation.
Structure, 24, 2016
|
|
6LPR
 
 | |
6VJR
 
 | | Oxygen tolerant Archeal 4hydroxybutyrylCoA dehydratase (4HBD) from N. maritimus | | Descriptor: | FE (III) ION, FLAVIN-ADENINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | DeMirci, H, Mathews, I.I, Wakatsuki, S, Kulakman, C. | | Deposit date: | 2020-01-17 | | Release date: | 2021-01-27 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural adaptation of oxygen tolerance in a key archaeal carbon fixation enzyme
To Be Published
|
|
6ZZA
 
 | | Crystal structure of CbpB in complex with c-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, CBS domain-containing protein, SULFATE ION | | Authors: | Mechaly, A.E, Covaleda-Cortes, G, Kaminski, P.A. | | Deposit date: | 2020-08-04 | | Release date: | 2021-08-18 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2.491 Å) | | Cite: | The c-di-AMP-binding protein CbpB modulates the level of ppGpp alarmone in Streptococcus agalactiae.
Febs J., 290, 2023
|
|
7FVU
 
 | | Crystal Structure of human FABP4 in complex with 2-[2-(benzothiophen-3-yl)-2,3-dihydrobenzothiophene-3-carbonyl]benzoic acid | | Descriptor: | 2-[(2S,3R)-2,3-dihydro[2,3'-bi-1-benzothiophene]-3-carbonyl]benzoic acid, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
