8WEG
 
 | | Crystal structure of Brassica napus MIK2 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wan, L.H, Hu, Y.X, Wu, H.M. | | Deposit date: | 2023-09-17 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Mechanistic study of SCOOPs recognition by MIK2-BAK1 complex reveals the role of N-glycans in plant ligand-receptor-coreceptor complex formation
Nature Plants, 2024
|
|
8WEH
 
 | | Crystal structure of Brassica napus MIK2 ectodomain (N393A mutant) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wan, L.H, Hu, Y.X, Wu, H.M. | | Deposit date: | 2023-09-17 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Mechanistic study of SCOOPs recognition by MIK2-BAK1 complex reveals the role of N-glycans in plant ligand-receptor-coreceptor complex formation
Nature Plants, 2024
|
|
8WEI
 
 | | Crystal structure of Brassica napus MIK2 ectodomain (N393D mutant) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wan, L.H, Hu, Y.X, Wu, H.M. | | Deposit date: | 2023-09-17 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Mechanistic study of SCOOPs recognition by MIK2-BAK1 complex reveals the role of N-glycans in plant ligand-receptor-coreceptor complex formation
Nature Plants, 2024
|
|
8WV1
 
 | |
8WVT
 
 | | Crystal structure of Lsd18 in complex with ligands | | Descriptor: | (4R,5S)-3-((2R,3S,4S,E)-2,6-diethyl-9-((2R,3R)-2-ethyl-3-methyloxiran-2-yl)-3-hydroxy-4-methylnon-6-enoyl)-4-methyl-5-phenyloxazolidin-2-one, (4R,5S)-4-methyl-5-phenyl-3-((2R,3S,4S,6E,10E)-2,6,10-triethyl-3-hydroxy-4-methyldodeca-6,10-dienoyl)oxazolidin-2-one, CHLORIDE ION, ... | | Authors: | Deng, Y.M, Hu, Y.L, Chen, X. | | Deposit date: | 2023-10-24 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of Lsd18 in complex with ligands
To Be Published
|
|
8WWG
 
 | |
8WXS
 
 | |
8WYW
 
 | |
8WZ6
 
 | | The crystal structure of Legionella pneumophila adenosylhomocysteinase Lpg2021 in ternary complex with NAD and DZNep | | Descriptor: | Adenosylhomocysteinase, DZNep, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Gao, Y.S, Xie, R, Chen, Y.N, Ma, J.M, Ge, H.H. | | Deposit date: | 2023-11-01 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | The crystal structure of Legionella pneumophila adenosylhomocysteinase Lpg2021 in ternary complex with NAD and DZNep
To Be Published
|
|
8WZ7
 
 | | The crystal structure of Legionella pneumophila adenosylhomocysteinase Lpg2021(I255A,T287A) in ternary complex with NAD and adenosine | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Gao, Y.S, Xie, R, Chen, Y.N, Ma, J.M, Ge, H.H. | | Deposit date: | 2023-11-01 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.922 Å) | | Cite: | Structural basis of substrate recognition for Legionella pneumophila SAH hydrolase Lpg2021
To Be Published
|
|
8WZ8
 
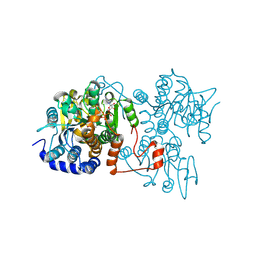 | | The crystal structure of Legionella pneumophila adenosylhomocysteinase Lpg2021(T67A,Q69A) complex with NAD | | Descriptor: | Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Gao, Y.S, Xie, R, Chen, Y.N, Ma, J.M, Ge, H.H. | | Deposit date: | 2023-11-01 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.583 Å) | | Cite: | The crystal structure of Legionella pneumophila adenosylhomocysteinase Lpg2021(T67A,Q69A) complex with NAD
To Be Published
|
|
8WZ9
 
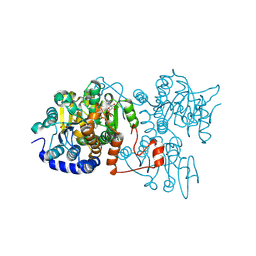 | | The crystal structure of Legionella pneumophila adenosylhomocysteinase Lpg2021 in ternary complex with NAD and adenine | | Descriptor: | ADENINE, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Gao, Y.S, Xie, R, Chen, Y.N, Ma, J.M, Ge, H.H. | | Deposit date: | 2023-11-01 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | The crystal structure of Legionella pneumophila adenosylhomocysteinase Lpg2021 in ternary complex with NAD and adenine
To Be Published
|
|
8WZZ
 
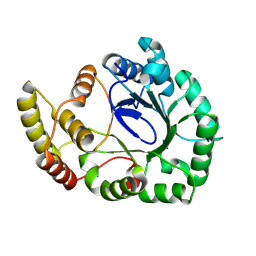 | |
8X0A
 
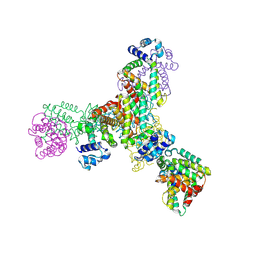 | |
8X0N
 
 | | The molecular mechanism of hnRNPA1 recognize TERRA RNA. | | Descriptor: | Heterogeneous nuclear ribonucleoprotein A1, RNA (5'-R(*UP*UP*AP*GP*GP*GP*UP*UP*AP*GP*GP*G)-3') | | Authors: | Xu, Z.Y, Liu, Y, Li, F. | | Deposit date: | 2023-11-05 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The molecular mechanism of hnRNPA1 recognize TERRA and regulate the telomeric stability through liquid-liquid phase separation.
To Be Published
|
|
8X18
 
 | | Xanthomonas campestris pv. campestris OpgD mutant-D379N with beta-1,2-glucan | | Descriptor: | GLYCEROL, Glucans biosynthesis protein D, beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose, ... | | Authors: | Motouchi, S, Nakajima, M. | | Deposit date: | 2023-11-06 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Xanthomonas campestris pv. campestris OpgD mutant-D379N with beta-1,2-glucan
To Be Published
|
|
8XM3
 
 | | Methionyl-tRNA synthetase from Staphylococcus aureus complexed with a chlorhexidine derivative and ATP | | Descriptor: | 1-(4-chlorophenyl)-3-[~{N}-[4-[[~{N}-[~{N}-(4-chlorophenyl)carbamimidoyl]carbamimidoyl]amino]butyl]carbamimidoyl]guanidine, ADENOSINE-5'-TRIPHOSPHATE, Methionine--tRNA ligase | | Authors: | Lu, F, Xia, K, Yi, J, Chen, B, Luo, Z, Xu, J, Gu, Q, Zhou, H. | | Deposit date: | 2023-12-27 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Biochemical and structural characterization of chlorhexidine as an ATP-assisted inhibitor against type 1 methionyl-tRNA synthetase from Gram-positive bacteria.
Eur.J.Med.Chem., 268, 2024
|
|
8XM4
 
 | | Methionyl-tRNA synthetase from Staphylococcus aureus in complex with chlorhexidine and ATP | | Descriptor: | 1,2-ETHANEDIOL, 1-[6-[azanylidene-[[azanylidene-[[(4-chlorophenyl)amino]methyl]-$l^{4}-azanyl]methyl]-$l^{4}-azanyl]hexyl]-3-[~{N}-(4-chlorophenyl)carbamimidoyl]guanidine, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Lu, F, Xia, K, Yi, J, Chen, B, Luo, Z, Xu, J, Gu, Q, Zhou, H. | | Deposit date: | 2023-12-27 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Biochemical and structural characterization of chlorhexidine as an ATP-assisted inhibitor against type 1 methionyl-tRNA synthetase from Gram-positive bacteria.
Eur.J.Med.Chem., 268, 2024
|
|
8XV4
 
 | |
8XV6
 
 | | Crystal structure of PHD domain of UHRF1 in complex with mStella peptide (residues 85-119) | | Descriptor: | Developmental pluripotency-associated protein 3, E3 ubiquitin-protein ligase UHRF1, GLYCEROL, ... | | Authors: | Du, X, Gan, Q, Xu, J, Liu, J. | | Deposit date: | 2024-01-14 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of PHD domain of UHRF1 in complex with mStella peptide (residues 85-119)
To Be Published
|
|
8XV7
 
 | | Crystal structure of TTD-PHD domain of UHRF1 in complex with hStella peptide (residues 75-121) | | Descriptor: | ACETIC ACID, Developmental pluripotency-associated protein 3, E3 ubiquitin-protein ligase UHRF1, ... | | Authors: | Du, X, Gan, Q, Xu, J, Liu, J. | | Deposit date: | 2024-01-14 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of TTD-PHD domain of UHRF1 in complex with hStella peptide (residues 75-121)
To Be Published
|
|
8XV8
 
 | |
8Y1Q
 
 | | Crystal structure of Aquifex aeolicus dUTPase complexed with dUMP. | | Descriptor: | 2'-DEOXYURIDINE-5'-MONOPHOSPHATE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, MAGNESIUM ION | | Authors: | Fukui, K, Murakawa, T, Yano, T. | | Deposit date: | 2024-01-25 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | dUTP pyrophosphatases from hyperthermophilic eubacterium and archaeon: Structural and functional examinations on the suitability for PCR application.
Protein Sci., 33, 2024
|
|
8Y1W
 
 | | Crystal structure of Thermococcus pacificus dUTPase complexed with dUMP. | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Fukui, K, Murakawa, T, Yano, T. | | Deposit date: | 2024-01-25 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | dUTP pyrophosphatases from hyperthermophilic eubacterium and archaeon: Structural and functional examinations on the suitability for PCR application.
Protein Sci., 33, 2024
|
|
8YWP
 
 | | Crystal structure of the Fab fragment of anti-IL-6 antibody I9H | | Descriptor: | GLYCEROL, Heavy chain of the Fab fragment of anti-IL-6 antibody I9H, Light chain of the Fab fragment of anti-IL-6 antibody I9H, ... | | Authors: | Yudenko, A, Bukhdruker, S, Eliseev, I, Rodin, S, Burtseva, A, Petrov, A, Zlobina, O, Ischenko, A, Borshchevskiy, V. | | Deposit date: | 2024-03-31 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of signaling complex inhibition by IL-6 domain-swapped dimers
Structure, 2025
|
|
