1NXT
 
 | | MicArec pH 4.0 | | Descriptor: | DNA-binding response regulator, XENON | | Authors: | Bent, C.J, Isaacs, N.W, Mitchell, T.J, Riboldi-Tunnicliffe, A. | | Deposit date: | 2003-02-11 | | Release date: | 2004-02-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal structure of the response regulator 02 receiver domain, the essential YycF two-component system of Streptococcus pneumoniae in both complexed and native states.
J.Bacteriol., 186, 2004
|
|
1NXU
 
 | | CRYSTAL STRUCTURE OF E. COLI HYPOTHETICAL OXIDOREDUCTASE YIAK NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ER82. | | Descriptor: | Hypothetical oxidoreductase yiaK, SULFATE ION | | Authors: | Forouhar, F, Lee, I, Benach, J, Kulkarni, K, Xiao, R, Acton, T.B, Shastry, R, Rost, B, Montelione, G.T, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-02-11 | | Release date: | 2003-03-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Novel NAD-binding Protein Revealed by the Crystal Structure of 2,3-Diketo-L-gulonate Reductase (YiaK).
J.Biol.Chem., 279, 2004
|
|
1NXV
 
 | |
1NXW
 
 | | MicArec pH 5.1 | | Descriptor: | ACETIC ACID, DNA-binding response regulator | | Authors: | Bent, C.J, Isaacs, N.W, Mitchell, T.J, Riboldi-Tunnicliffe, A. | | Deposit date: | 2003-02-11 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of the response regulator 02 receiver domain, the essential YycF two-component system of Streptococcus pneumoniae in both complexed and native states.
J.Bacteriol., 186, 2004
|
|
1NXX
 
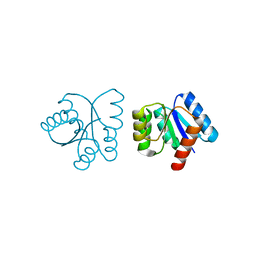 | |
1NXY
 
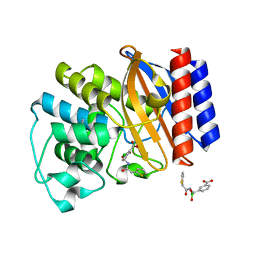 | | Crystal Structure of the complex between M182T mutant of TEM-1 and a boronic acid inhibitor (SM2) | | Descriptor: | (1R)-1-(2-THIENYLACETYLAMINO)-1-(3-CARBOXYPHENYL)METHYLBORONIC ACID, Beta-lactamase TEM, POTASSIUM ION | | Authors: | Wang, X, Minasov, G, Blazquez, J, Caselli, E, Prati, F, Shoichet, B.K. | | Deposit date: | 2003-02-11 | | Release date: | 2003-08-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Recognition and Resistance in TEM beta-lactamase
Biochemistry, 42, 2003
|
|
1NXZ
 
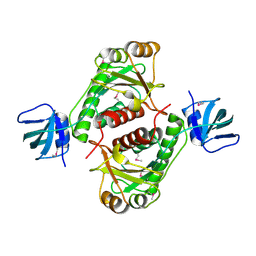 | | X-Ray Crystal Structure of Protein yggj_haein of Haemophilus influenzae. Northeast Structural Genomics Consortium Target IR73. | | Descriptor: | Hypothetical protein HI0303 | | Authors: | Forouhar, F, Shen, J, Xiao, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-02-11 | | Release date: | 2003-03-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional assignment based on structural analysis: crystal structure of the yggJ protein (HI0303) of Haemophilus influenzae reveals an RNA methyltransferase with a deep trefoil knot.
Proteins, 53, 2003
|
|
1NY0
 
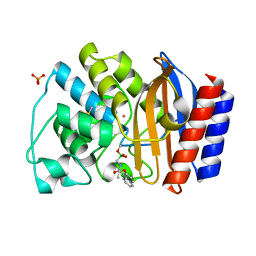 | | Crystal Structure of the complex between M182T mutant of TEM-1 and a boronic acid inhibitor (NBF) | | Descriptor: | Beta-lactamase TEM, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Wang, X, Minasov, G, Blazquez, J, Caselli, E, Prati, F, Shoichet, B.K. | | Deposit date: | 2003-02-11 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Recognition and Resistance in TEM beta-lactamase
Biochemistry, 42, 2003
|
|
1NY1
 
 | | CRYSTAL STRUCTURE OF B. SUBTILIS POLYSACCHARIDE DEACETYLASE NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET SR127. | | Descriptor: | Probable polysaccharide deacetylase pdaA | | Authors: | Forouhar, F, Edstrom, W, Khan, J, Ma, L, Chiang, Y, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-02-11 | | Release date: | 2003-03-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Polysaccharide Deacetylase (PDAA_BACSU) from B. Subtilis (Pdaa_Bacsu) Northeast Structural Genomics Research Consortium (Nesg) Target Sr127
To be Published
|
|
1NY5
 
 | | Crystal structure of sigm54 activator (AAA+ ATPase) in the inactive state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Lee, S.Y, de la Torre, A, Kustu, S, Nixon, B.T, Wemmer, D.E. | | Deposit date: | 2003-02-11 | | Release date: | 2003-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Regulation of the transcriptional activator NtrC1: structural studies of the regulatory and AAA+ ATPase domains
Genes Dev., 17, 2003
|
|
1NY6
 
 | | Crystal structure of sigm54 activator (AAA+ ATPase) in the active state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, transcriptional regulator (NtrC family) | | Authors: | Lee, S.Y, de la Torre, A, Kustu, S, Nixon, B.T, Wemmer, D.E. | | Deposit date: | 2003-02-11 | | Release date: | 2003-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Regulation of the transcriptional activator NtrC1: structural studies of the regulatory and AAA+ ATPase domains
Genes Dev., 17, 2003
|
|
1NY7
 
 | | COWPEA MOSAIC VIRUS (CPMV) | | Descriptor: | COWPEA MOSAIC VIRUS, LARGE (L) SUBUNIT, SMALL (S) SUBUNIT | | Authors: | Lin, T, Chen, Z, Usha, R, Stauffacher, C.V, Dai, J.-B, Schmidt, T, Johnson, J.E. | | Deposit date: | 2003-02-11 | | Release date: | 2003-03-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Refined Crystal Structure of Cowpea Mosaic Virus at 2.8A Resolution
Virology, 265, 1999
|
|
1NYC
 
 | | Staphostatins resemble lipocalins, not cystatins in fold. | | Descriptor: | CHLORIDE ION, SULFATE ION, cysteine protease inhibitor | | Authors: | Rzychon, M, Filipek, R, Sabat, A, Kosowska, K, Dubin, A, Potempa, J, Bochtler, M. | | Deposit date: | 2003-02-12 | | Release date: | 2003-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Staphostatins resemble lipocalins, not cystatins in fold.
Protein Sci., 12, 2003
|
|
1NYE
 
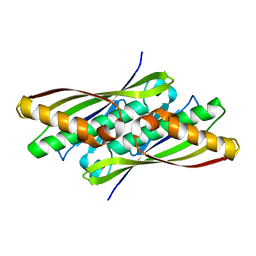 | | Crystal structure of OsmC from E. coli | | Descriptor: | Osmotically inducible protein C | | Authors: | Shin, D.H, Choi, I.-G, Busso, D, Jancarik, J, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2003-02-12 | | Release date: | 2004-03-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of OsmC from Escherichia coli: a salt-shock-induced protein.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1NYH
 
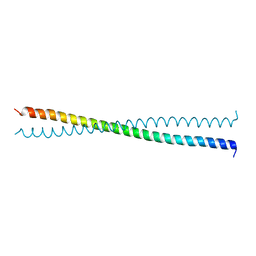 | | Crystal Structure of the Coiled-coil Dimerization Motif of Sir4 | | Descriptor: | Regulatory protein SIR4 | | Authors: | Chang, J.F, Hall, B.E, Tanny, J.C, Moazed, D, Filman, D, Ellenberger, T. | | Deposit date: | 2003-02-12 | | Release date: | 2003-06-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the Coiled-coil Dimerization Motif of Sir4 and Its Interaction With Sir3
Structure, 11, 2003
|
|
1NYK
 
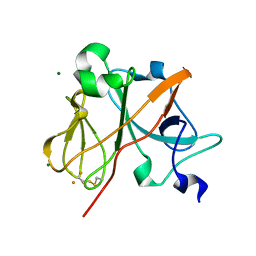 | | Crystal Structure of the Rieske protein from Thermus thermophilus | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, MAGNESIUM ION, Rieske iron-sulfur protein | | Authors: | Hunsicker-Wang, L.M, Heine, A, Chen, Y, Luna, E.P, Todaro, T, Zhang, Y.M, Williams, P.A, McRee, D.E, Hirst, J, Stout, C.D, Fee, J.A. | | Deposit date: | 2003-02-12 | | Release date: | 2003-06-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | High resolution structure of the soluble, respiratory-type Rieske protein from Thermus thermophilus: Analysis and Comparison
Biochemistry, 42, 2003
|
|
1NYL
 
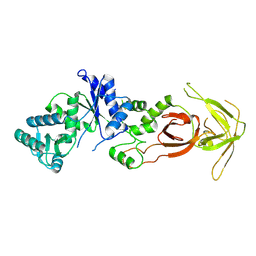 | | Unliganded glutaminyl-tRNA synthetase | | Descriptor: | Glutaminyl-tRNA synthetase | | Authors: | Sherlin, L.D, Perona, J.P. | | Deposit date: | 2003-02-12 | | Release date: | 2003-02-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | tRNA-Dependent Active Site Assembly in a Class I Aminoacyl-tRNA Synthetase.
Structure, 11, 2003
|
|
1NYM
 
 | | Crystal Structure of the complex between M182T mutant of TEM-1 and a boronic acid inhibitor (CXB) | | Descriptor: | Beta-lactamase TEM, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Wang, X, Minasov, G, Blazquez, J, Caselli, E, Prati, F, Shoichet, B.K. | | Deposit date: | 2003-02-12 | | Release date: | 2003-08-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Recognition and resistance in TEM beta-lactamase
Biochemistry, 42, 2003
|
|
1NYQ
 
 | | Structure of Staphylococcus aureus threonyl-tRNA synthetase complexed with an analogue of threonyl adenylate | | Descriptor: | 5'-O-(N-(L-THREONYL)-SULFAMOYL)ADENOSINE, ZINC ION, threonyl-tRNA synthetase 1 | | Authors: | Torres-Larios, A, Sankaranarayanan, R, Rees, B, Dock-Bregeon, A.C, Moras, D. | | Deposit date: | 2003-02-13 | | Release date: | 2003-10-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Conformational movements and cooperativity upon amino acid, ATP and tRNA binding in threonyl-tRNA synthetase
J.Mol.Biol., 331, 2003
|
|
1NYR
 
 | | Structure of Staphylococcus aureus threonyl-tRNA synthetase complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, THREONINE, ZINC ION, ... | | Authors: | Torres-Larios, A, Sankaranarayanan, R, Rees, B, Dock-Bregeon, A.C, Moras, D. | | Deposit date: | 2003-02-13 | | Release date: | 2003-10-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational movements and cooperativity upon amino acid, ATP and tRNA binding in threonyl-tRNA synthetase
J.Mol.Biol., 331, 2003
|
|
1NYS
 
 | |
1NYT
 
 | | SHIKIMATE DEHYDROGENASE AroE COMPLEXED WITH NADP+ | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION, ... | | Authors: | Roszak, A.W, Lapthorn, A.J. | | Deposit date: | 2003-02-13 | | Release date: | 2003-03-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of shikimate dehydrogenase AroE and its Paralog YdiB. A common structural framework for different activities.
J.Biol.Chem., 278, 2003
|
|
1NYU
 
 | |
1NYW
 
 | | The high resolution structures of RmlC from Streptoccus suis in complex with dTDP-D-glucose | | Descriptor: | 2'DEOXY-THYMIDINE-5'-DIPHOSPHO-ALPHA-D-GLUCOSE, dTDP-6-deoxy-D-xylo-4-hexulose 3,5-epimerase | | Authors: | Dong, C, Major, L.L, Allen, A, Blankenfeldt, W, Maskell, D, Naismith, J.H. | | Deposit date: | 2003-02-14 | | Release date: | 2003-06-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-Resolution Structures of RmlC from Streptococcus suis in Complex with Substrate Analogs Locate the Active Site of This Class of Enzyme
Structure, 11, 2003
|
|
1NYY
 
 | | Crystal Structure of the complex between M182T mutant of TEM-1 and a boronic acid inhibitor (105) | | Descriptor: | Beta-lactamase TEM, N-[5-METHYL-3-O-TOLYL-ISOXAZOLE-4-CARBOXYLIC ACID AMIDE] BORONIC ACID | | Authors: | Wang, X, Minasov, G, Blazquez, J, Caselli, E, Prati, F, Shoichet, B.K. | | Deposit date: | 2003-02-14 | | Release date: | 2003-08-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Recognition and resistance in TEM beta-lactamase
Biochemistry, 42, 2003
|
|
