1K2C
 
 | | Combining Mutations in HIV-1 Protease to Understand Mechanisms of Resistance | | Descriptor: | N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide, PROTEASE RETROPEPSIN | | Authors: | Mahalingam, B, Boross, P, Wang, Y.-F, Louis, J.M, Fischer, C, Tozser, J, W Harrison, R, Weber, I.T. | | Deposit date: | 2001-09-26 | | Release date: | 2002-07-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Combining mutations in HIV-1 protease to understand mechanisms of resistance.
Proteins, 48, 2002
|
|
1K2E
 
 | | crystal structure of a nudix protein from Pyrobaculum aerophilum | | Descriptor: | ACETIC ACID, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Wang, S, Mura, C, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2001-09-26 | | Release date: | 2002-04-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a Nudix protein from Pyrobaculum aerophilum reveals a dimer with two intersubunit beta-sheets.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1K2F
 
 | | siah, Seven In Absentia Homolog | | Descriptor: | BETA-MERCAPTOETHANOL, ZINC ION, siah-1A protein | | Authors: | Polekhina, G, House, C.M, Traficante, N, Mackay, J.P, Relaix, F, Sassoon, D.A, Parker, M.W, Bowtell, D.D.L. | | Deposit date: | 2001-09-26 | | Release date: | 2001-12-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Siah ubiquitin ligase is structurally related to TRAF and modulates TNF-alpha signaling.
Nat.Struct.Biol., 9, 2002
|
|
1K2I
 
 | | Crystal Structure of Gamma-Chymotrypsin in Complex with 7-Hydroxycoumarin | | Descriptor: | 2,4-DIHYDROXY-TRANS CINNAMIC ACID, CHYMOTRYPSINOGEN A, SULFATE ION | | Authors: | Ghani, U, Ng, K.K.S, Atta-ur-Rahman, Choudhary, M.I, Ullah, N, James, M.N.G. | | Deposit date: | 2001-09-27 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of gamma-chymotrypsin in complex with 7-hydroxycoumarin.
J.Mol.Biol., 314, 2001
|
|
1K2L
 
 | | STRUCTURAL CHARACTERIZATION OF BISINTERCALATION IN HIGHER-ORDER DNA AT A JUNCTION-LIKE QUADRUPLEX | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*G)-3', BIS-(9-OCTYLAMINO(2-DIMETHYLAMINOETHYL)ACRIDINE-4-CARBOXAMIDE, COBALT (II) ION | | Authors: | Teixeira, S.C.M, Thorpe, J.H, Todd, A.K, Powell, H.R, Adams, A, Wakelin, L.P.G, Denny, W.A, Cardin, C.J. | | Deposit date: | 2001-09-28 | | Release date: | 2002-10-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Characterisation of Bisintercalation in Higher-order DNA at a Junction-like Quadruplex
J.MOL.BIOL., 323, 2002
|
|
1K2R
 
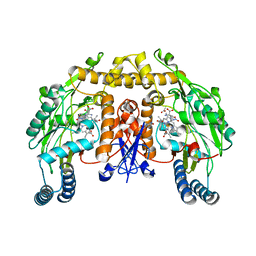 | | Structure of rat brain nNOS heme domain complexed with NG-nitro-L-arginine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-OMEGA-NITRO-L-ARGININE, ... | | Authors: | Li, H, Martasek, P, Masters, B.S.S, Poulos, T.L, Raman, C.S. | | Deposit date: | 2001-09-28 | | Release date: | 2003-03-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of rat brain nNOS heme domain
To be Published
|
|
1K2S
 
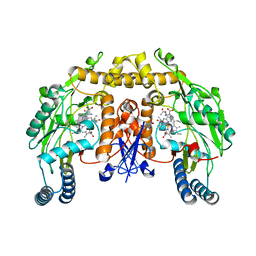 | | Structure of rat brain nNOS heme domain complexed with NG-allyl-L-arginine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 5-N-ALLYL-ARGININE, ACETATE ION, ... | | Authors: | Li, H, Martasek, P, Masters, B.S.S, Poulos, T.L, Raman, C.S. | | Deposit date: | 2001-09-28 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of rat brain nNOS heme domain
To be Published
|
|
1K2T
 
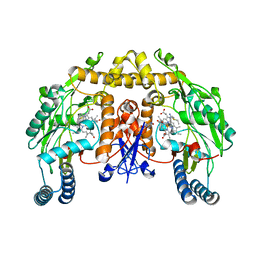 | | Structure of rat brain nNOS heme domain complexed with S-ethyl-N-phenyl-isothiourea | | Descriptor: | 2-ETHYL-1-PHENYL-ISOTHIOUREA, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Martasek, P, Masters, B.S.S, Poulos, T.L, Raman, C.S. | | Deposit date: | 2001-09-28 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of rat brain nNOS heme domain
To be Published
|
|
1K2U
 
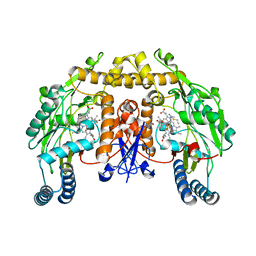 | | Structure of rat brain nNOS heme domain complexed with S-ethyl-N-[4-(trifluoromethyl)phenyl] isothiourea | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Li, H, Martasek, P, Masters, B.S.S, Poulos, T.L, Raman, C.S. | | Deposit date: | 2001-09-28 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of rat brain nNOS heme domain
To be Published
|
|
1K2W
 
 | |
1K2X
 
 | |
1K2Z
 
 | |
1K30
 
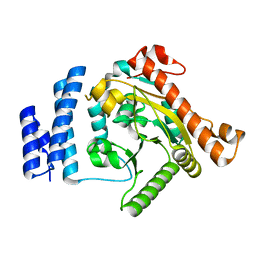 | | Crystal Structure Analysis of Squash (Cucurbita moschata) glycerol-3-phosphate (1)-acyltransferase | | Descriptor: | glycerol-3-phosphate acyltransferase | | Authors: | Turnbull, A.P, Rafferty, J.B, Sedelnikova, S.E, Slabas, A.R, Schierer, T.P, Kroon, J.T, Simon, J.W, Fawcett, T, Nishida, I, Murata, N, Rice, D.W. | | Deposit date: | 2001-10-01 | | Release date: | 2001-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Analysis of the structure, substrate specificity, and mechanism of squash glycerol-3-phosphate (1)-acyltransferase.
Structure, 9, 2001
|
|
1K32
 
 | |
1K33
 
 | | Crystal structure analysis of the gp41 core mutant | | Descriptor: | Transmembrane glycoprotein GP41 | | Authors: | Shu, W, Lu, M. | | Deposit date: | 2001-10-01 | | Release date: | 2001-10-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Interhelical interactions in the gp41 core: implications for activation of HIV-1 membrane fusion.
Biochemistry, 41, 2002
|
|
1K34
 
 | | Crystal structure analysis of gp41 core mutant | | Descriptor: | Transmembrane glycoprotein GP41 | | Authors: | Shu, W, Lu, M. | | Deposit date: | 2001-10-01 | | Release date: | 2001-10-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Interhelical interactions in the gp41 core: implications for activation of HIV-1 membrane fusion.
Biochemistry, 41, 2002
|
|
1K38
 
 | | CRYSTAL STRUCTURE OF THE CLASS D BETA-LACTAMASE OXA-2 | | Descriptor: | Beta-lactamase OXA-2, FORMIC ACID | | Authors: | Kerff, F, Fonze, E, Bouillenne, F, Frere, J.M, Charlier, P. | | Deposit date: | 2001-10-02 | | Release date: | 2003-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | CRYSTAL STRUCTURE OF THE CLASS D BETA-LACTAMASE OXA-2
To be Published
|
|
1K39
 
 | | The structure of yeast delta3-delta2-enoyl-COA isomerase complexed with octanoyl-COA | | Descriptor: | OCTANOYL-COENZYME A, PHOSPHATE ION, d3,d2-enoyl CoA isomerase ECI1 | | Authors: | Mursula, A.M, Geerlof, A, Hiltunen, J.K, Wierenga, R.K. | | Deposit date: | 2001-10-02 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: |
|
|
1K3A
 
 | | Structure of the Insulin-like Growth Factor 1 Receptor Kinase | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, insulin receptor substrate 1, insulin-like growth factor 1 receptor | | Authors: | Favelyukis, S, Till, J.H, Hubbard, S.R, Miller, W.T. | | Deposit date: | 2001-10-02 | | Release date: | 2001-11-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and autoregulation of the insulin-like growth factor 1 receptor kinase.
Nat.Struct.Biol., 8, 2001
|
|
1K3B
 
 | | Crystal Structure of Human Dipeptidyl Peptidase I (Cathepsin C): Exclusion Domain Added to an Endopeptidase Framework Creates the Machine for Activation of Granular Serine Proteases | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, SULFATE ION, ... | | Authors: | Turk, D, Janjic, V, Stern, I, Podobnik, M, Lamba, D, Dahl, S.W, Lauritzen, C, Pedersen, J, Turk, V, Turk, B. | | Deposit date: | 2001-10-02 | | Release date: | 2002-04-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of human dipeptidyl peptidase I (cathepsin C): exclusion domain added to an endopeptidase framework creates the machine for activation of granular serine proteases.
EMBO J., 20, 2001
|
|
1K3C
 
 | | Phosphoenolpyruvate carboxykinase in complex with ADP, AlF3 and Pyruvate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Sudom, A.M, Prasad, L, Goldie, H, Delbaere, L.T.J. | | Deposit date: | 2001-10-02 | | Release date: | 2001-12-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The phosphoryl-transfer mechanism of Escherichia coli phosphoenolpyruvate carboxykinase from the use of AlF(3).
J.Mol.Biol., 314, 2001
|
|
1K3D
 
 | | Phosphoenolpyruvate carboxykinase in complex with ADP and AlF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Sudom, A.M, Prasad, L, Goldie, H, Delbaere, L.T.J. | | Deposit date: | 2001-10-02 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The phosphoryl-transfer mechanism of Escherichia coli phosphoenolpyruvate carboxykinase from the use of AlF(3).
J.Mol.Biol., 314, 2001
|
|
1K3E
 
 | | Type III secretion chaperone CesT | | Descriptor: | CesT | | Authors: | Luo, Y, Bertero, M, Frey, E.A, Pfuetzner, R.A, Wenk, M.R, Creagh, L, Marcus, S.L, Lim, D, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2001-10-02 | | Release date: | 2001-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and biochemical characterization of the type III secretion chaperones CesT and SigE.
Nat.Struct.Biol., 8, 2001
|
|
1K3I
 
 | | Crystal Structure of the Precursor of Galactose Oxidase | | Descriptor: | ACETATE ION, CALCIUM ION, Galactose Oxidase Precursor, ... | | Authors: | Firbank, S.J, Rogers, M.S, Wilmot, C.M, Dooley, D.M, Halcrow, M.A, Knowles, P.F, McPherson, M.J, Phillips, S.E.V. | | Deposit date: | 2001-10-03 | | Release date: | 2001-11-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the precursor of galactose oxidase: an unusual self-processing enzyme.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1K3L
 
 | | Crystal Structure Analysis of S-hexyl-glutathione Complex of Glutathione Transferase at 1.5 Angstroms Resolution | | Descriptor: | GLUTATHIONE S-TRANSFERASE A1, S-HEXYLGLUTATHIONE | | Authors: | Le Trong, I, Stenkamp, R.E, Ibarra, C, Atkins, W.M, Adman, E.T. | | Deposit date: | 2001-10-03 | | Release date: | 2002-10-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.3-A resolution structure of human glutathione S-transferase with S-hexyl
glutathione bound reveals possible extended ligandin binding site
Proteins, 48, 2002
|
|
