1N50
 
 | |
1N51
 
 | | Aminopeptidase P in complex with the inhibitor apstatin | | Descriptor: | MANGANESE (II) ION, Xaa-Pro aminopeptidase, apstatin | | Authors: | Graham, S.C, Maher, M.J, Lee, M.H, Simmons, W.H, Freeman, H.C, Guss, J.M. | | Deposit date: | 2002-11-03 | | Release date: | 2003-12-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Escherichia coli aminopeptidase P in complex with the inhibitor apstatin.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1N52
 
 | | Cap Binding Complex | | Descriptor: | 20 kDa nuclear cap binding protein, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, 80 kDa nuclear cap binding protein, ... | | Authors: | Calero, G, Wilson, K, Ly, T, Rios-Steiner, J, Clardy, J, Cerione, R. | | Deposit date: | 2002-11-04 | | Release date: | 2003-02-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural basis of m7GpppG binding to the nuclear cap-binding protein complex.
Nat.Struct.Biol., 9, 2002
|
|
1N54
 
 | | Cap Binding Complex m7GpppG free | | Descriptor: | 20 kDa nuclear cap binding protein, 80 kDa nuclear cap binding protein, GLYCEROL | | Authors: | Calero, G, Wilson, K, Ly, T, Rios-Steiner, J, Clardy, J, Cerione, R. | | Deposit date: | 2002-11-04 | | Release date: | 2003-02-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural basis of m7GpppG binding to the nuclear cap-binding protein complex.
Nat.Struct.Biol., 9, 2002
|
|
1N55
 
 | |
1N56
 
 | | Y-family DNA polymerase Dpo4 in complex with DNA containing abasic lesion | | Descriptor: | 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*CP*TP*AP*A)-3', 5'-D(*TP*CP*AP*TP*(3DR)P*AP*GP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3', ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Ling, H, Boudsocq, F, Woodgate, R, Yang, W. | | Deposit date: | 2002-11-04 | | Release date: | 2004-02-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Snapshots of replication through an abasic lesion; structural basis for base substitutions and frameshifts.
Mol.Cell, 13, 2004
|
|
1N57
 
 | | Crystal Structure of Chaperone Hsp31 | | Descriptor: | Chaperone Hsp31, MAGNESIUM ION | | Authors: | Quigley, P.M, Korotkov, K, Baneyx, F, Hol, W.G.J. | | Deposit date: | 2002-11-04 | | Release date: | 2003-03-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The 1.6A Crystal Structure of the Class of Chaperone Represented by
Escherichia coli Hsp31 Reveals a Putative Catalytic Triad
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1N59
 
 | | Crystal structure of the Murine class I Major Histocompatibility Complex of H-2KB, B2-Microglobulin, and A 9-Residue immunodominant peptide epitope gp33 derived from LCMV | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, K-B alpha chain, ... | | Authors: | Achour, A, Michaelsson, J, Harris, R.A, Odeberg, J, Grufman, P, Sandberg, J.K, Levitsky, V, Karre, K, Sandalova, T, Schneider, G. | | Deposit date: | 2002-11-05 | | Release date: | 2003-01-07 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | A Structural Basis for LCMV Immune Evasion. Subversion of H-2D(b) and H-2K(b) Presentation of gp33
Revealed by Comparative Crystal Structure Analyses.
Immunity, 17, 2002
|
|
1N5A
 
 | | Crystal structure of the Murine class I Major Histocompatibility Complex of H-2DB, B2-Microglobulin, and A 9-Residue immunodominant peptide epitope gp33 derived from LCMV | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Achour, A, Michaelsson, J, Harris, R.A, Odeberg, J, Grufman, P, Sandberg, J.K, Levitsky, V, Karre, K, Sandalova, T, Schneider, G. | | Deposit date: | 2002-11-05 | | Release date: | 2003-01-07 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A Structural Basis for LCMV Immune Evasion. Subversion of H-2D(b) and H-2K(b) Presentation of
gp33 Revealed by Comparative Crystal Structure Analyses.
Immunity, 17, 2002
|
|
1N5B
 
 | |
1N5C
 
 | | Crystal Structure Analysis of the B-DNA Dodecamer CGCGAATT(ethenoC)GCG | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(EDC)P*GP*CP*G)-3', SODIUM ION | | Authors: | Freisinger, E, Fernandes, A, Grollman, A.P, Kisker, C.F. | | Deposit date: | 2002-11-05 | | Release date: | 2003-06-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystallographic Characterization of an Exocyclic DNA Adduct: 3,N4-etheno-2'-deoxycytidine in the Dodecamer 5'-CGCGAATT(ethenoC)GCG-3'
J.Mol.Biol., 329, 2003
|
|
1N5I
 
 | | CRYSTAL STRUCTURE OF INACTIVE MYCOBACTERIUM TUBERCULOSIS THYMIDYLATE KINASE COMPLEXED WITH THYMIDINE MONOPHOSPHATE (TMP) AT PH 4.6 (RESOLUTION 1.85 A) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CITRATE ANION, SULFATE ION, ... | | Authors: | Fioravanti, E, Haouz, A, Ursby, T, Munier-Lehmann, H, Delarue, M, Bourgeois, D. | | Deposit date: | 2002-11-06 | | Release date: | 2003-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mycobacterium tuberculosis Thymidylate Kinase: Structural Studies of Intermediates along the Reaction Pathway
J.Mol.Biol., 327, 2003
|
|
1N5J
 
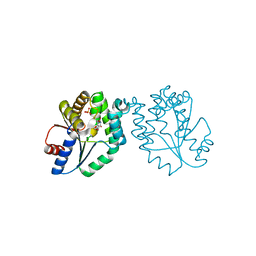 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS THYMIDYLATE KINASE COMPLEXED WITH THYMIDINE DIPHOSPHATE (TDP) AND THYMIDINE TRIPHOSPHATE (TTP) AT PH 5.4 (1.85 A RESOLUTION) | | Descriptor: | MAGNESIUM ION, SULFATE ION, THYMIDINE-5'-DIPHOSPHATE, ... | | Authors: | Fioravanti, E, Haouz, A, Ursby, T, Munier-Lehmann, H, Delarue, M, Bourgeois, D. | | Deposit date: | 2002-11-06 | | Release date: | 2003-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mycobacterium tuberculosis Thymidylate Kinase: Structural Studies of Intermediates along the
Reaction Pathway
J.Mol.Biol., 327, 2003
|
|
1N5K
 
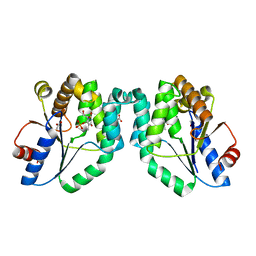 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS THYMIDYLATE KINASE CRYSTALLIZED IN SODIUM MALONATE (RESOLUTION 2.1 A) | | Descriptor: | ACETATE ION, MAGNESIUM ION, THYMIDINE-5'-PHOSPHATE, ... | | Authors: | Fioravanti, E, Haouz, A, Ursby, T, Munier-Lehmann, H, Delarue, M, Bourgeois, D. | | Deposit date: | 2002-11-06 | | Release date: | 2003-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mycobacterium tuberculosis Thymidylate Kinase: Structural Studies of Intermediates along the
Reaction Pathway
J.Mol.Biol., 375, 2003
|
|
1N5L
 
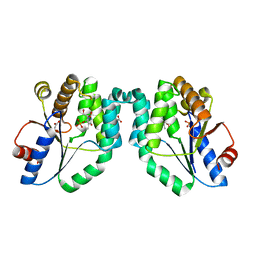 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS THYMIDYLATE KINASE CRYSTALLIZED IN SODIUM MALONATE, AFTER CATALYSIS IN THE CRYSTAL (2.3 A RESOLUTION) | | Descriptor: | ACETATE ION, DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Fioravanti, E, Haouz, A, Ursby, T, Munier-Lehmann, H, Delarue, M, Bourgeois, D. | | Deposit date: | 2002-11-06 | | Release date: | 2003-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mycobacterium tuberculosis Thymidylate Kinase: Structural Studies of Intermediates along the
Reaction Pathway
J.Mol.Biol., 375, 2003
|
|
1N5M
 
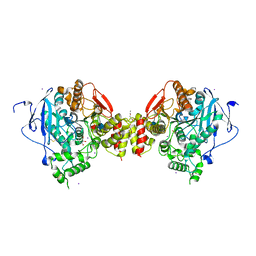 | | Crystal structure of the mouse acetylcholinesterase-gallamine complex | | Descriptor: | 2,2',2"-[1,2,3-BENZENE-TRIYLTRIS(OXY)]TRIS[N,N,N-TRIETHYLETHANAMINIUM], 2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, ... | | Authors: | Bourne, Y, Taylor, P, Radic, Z, Marchot, P. | | Deposit date: | 2002-11-06 | | Release date: | 2003-02-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into ligand interactions at the acetylcholinesterase peripheral anionic site
EMBO J., 22, 2003
|
|
1N5O
 
 | |
1N5Q
 
 | | Crystal structure of a Monooxygenase from the gene ActVA-Orf6 of Streptomyces coelicolor in complex with dehydrated Sancycline | | Descriptor: | 4-DIMETHYLAMINO-1,10,11,12-TETRAHYDROXY-3-OXO-3,4,4A,5-TETRAHYDRO-NAPHTHACENE-2-CARBOXYLIC ACID AMIDE, ActaVA-Orf6 monooxygenase, HEXAETHYLENE GLYCOL | | Authors: | Sciara, G, Kendrew, S.G, Miele, A.E, Marsh, N.G, Federici, L, Malatesta, F, Schimperna, G, Savino, C, Vallone, B. | | Deposit date: | 2002-11-07 | | Release date: | 2003-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The structure of ActVA-Orf6, a novel type of monooxygenase involved in actinorhodin biosynthesis
Embo J., 22, 2003
|
|
1N5R
 
 | | Crystal structure of the mouse acetylcholinesterase-propidium complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,8-DIAMINO-5[3-(DIETHYLMETHYLAMMONIO)PROPYL]-6-PHENYLPHENANTHRIDINIUM, ACETIC ACID, ... | | Authors: | Bourne, Y, Taylor, P, Radic, Z, Marchot, P. | | Deposit date: | 2002-11-07 | | Release date: | 2003-02-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights into ligand interactions at the acetylcholinesterase peripheral anionic site
EMBO J., 22, 2003
|
|
1N5S
 
 | | Crystal structure of a Monooxygenase from the gene ActVA-Orf6 of Streptomyces coelicolor in complex with the ligand Acetyl Dithranol | | Descriptor: | (1,8-DIHYDROXY-9-OXO-9,10-DIHYDRO-ANTHRACEN-2-YL)-ACETIC ACID, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, ActVA-Orf6 monooxygenase | | Authors: | Sciara, G, Kendrew, S.G, Miele, A.E, Marsh, N.G, Federici, L, Malatesta, F, Schimperna, G, Savino, C, Vallone, B. | | Deposit date: | 2002-11-07 | | Release date: | 2003-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of ActVA-Orf6, a novel type of monooxygenase involved in
actinorhodin biosynthesis
Embo J., 22, 2003
|
|
1N5T
 
 | | Crystal structure of a Monooxygenase from the gene ActVA-Orf6 of Streptomyces coelicolor in complex with the ligand Oxidized Acetyl Dithranol | | Descriptor: | (1,8-DIHYDROXY-9,10-DIOXO-9,10-DIHYDRO-ANTHRACEN-2-YL)-ACETIC ACID, ActVA-Orf6 monooxygenase | | Authors: | Sciara, G, G Kendrew, S, Miele, A.E, Marsh, N.G, Federici, L, Malatesta, F, Schimperna, G, Savino, C, Vallone, B. | | Deposit date: | 2002-11-07 | | Release date: | 2003-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of ActVA-Orf6, a novel type of monooxygenase involved in
actinorhodin biosynthesis
Embo J., 22, 2003
|
|
1N5U
 
 | | X-RAY STUDY OF HUMAN SERUM ALBUMIN COMPLEXED WITH HEME | | Descriptor: | MYRISTIC ACID, PROTOPORPHYRIN IX CONTAINING FE, SERUM ALBUMIN | | Authors: | Wardell, M, Wang, Z, Ho, J.X, Robert, J, Ruker, F, Ruble, J, Carter, D.C. | | Deposit date: | 2002-11-07 | | Release date: | 2003-06-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Atomic Structure of Human Methemalbumin at 1.9 A
Biochem.Biophys.Res.Commun., 291, 2002
|
|
1N5V
 
 | | Crystal structure of a Monooxygenase from the gene ActVA-Orf6 of Streptomyces coelicolor in complex with the ligand Nanaomycin D | | Descriptor: | 7-HYDROXY-5-METHYL-3,3A,5,11B-TETRAHYDRO-1,4-DIOXA-CYCLOPENTA[A]ANTHRACENE-2,6,11-TRIONE, ActVA-Orf6 monooxygenase | | Authors: | Sciara, G, Kendrew, S.G, Miele, A.E, Marsh, N.G, Federici, L, Malatesta, F, Schimperna, G, Savino, C, Vallone, B. | | Deposit date: | 2002-11-07 | | Release date: | 2003-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | The structure of ActVA-Orf6, a novel type of monooxygenase involved in
actinorhodin biosynthesis
Embo J., 22, 2003
|
|
1N5Y
 
 | | HIV-1 Reverse Transcriptase Crosslinked to Post-Translocation AZTMP-Terminated DNA (Complex P) | | Descriptor: | 5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(ATM))-3', 5'-D(*AP*TP*GP*C*TP*AP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3', MAGNESIUM ION, ... | | Authors: | Sarafianos, S.G, Clark Jr, A.D, Das, K, Tuske, S, Birktoft, J.J, Ilankumaran, P, Ramesha, A.R, Sayer, J.M, Jerina, D.M, Boyer, P.L, Hughes, S.H, Arnold, E. | | Deposit date: | 2002-11-07 | | Release date: | 2003-01-28 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of HIV-1 Reverse Transcriptase with Pre-Translocation and Post-Translocation AZTMP-Terminated DNA
Embo J., 21, 2002
|
|
1N5Z
 
 | | Complex structure of Pex13p SH3 domain with a peptide of Pex14p | | Descriptor: | 14-mer peptide from Peroxisomal membrane protein PEX14, Peroxisomal membrane protein PAS20 | | Authors: | Douangamath, A, Filipp, F.V, Klein, A.T.J, Barnett, P, Zou, P, Voorn-Brouwer, T, Vega, M.C, Mayans, O.M, Sattler, M, Distel, B, Wilmanns, M. | | Deposit date: | 2002-11-08 | | Release date: | 2002-12-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Topography for Independent Binding of alpha-Helical and PPII-Helical Ligands to a Peroxisomal SH3 Domain
MOL.CELL, 10, 2002
|
|
