1M4A
 
 | | Crystal Structure of Human Interleukin-2 Y31C Covalently Modified at C31 with (1H-Indol-3-yl)-(2-mercapto-ethoxyimino)-acetic acid | | Descriptor: | (1H-INDOL-3-YL)-(2-MERCAPTO-ETHOXYIMINO)-ACETIC ACID, GLYCEROL, interleukin-2 | | Authors: | Arkin, M.A, Randal, M, DeLano, W.L, Hyde, J, Luong, T.N, Oslob, J.D, Raphael, D.R, Taylor, L, Wang, J, McDowell, R.S, Wells, J.A, Braisted, A.C. | | Deposit date: | 2002-07-02 | | Release date: | 2002-07-31 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Binding of small molecules to an adaptive
protein-protein interface
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1M4B
 
 | | Crystal Structure of Human Interleukin-2 K43C Covalently Modified at C43 with 2-[2-(2-Cyclohexyl-2-guanidino-acetylamino)-acetylamino]-N-(3-mercapto-propyl)-propionamide | | Descriptor: | 2-[2-(2-CYCLOHEXYL-2-GUANIDINO-ACETYLAMINO)-ACETYLAMINO]-N-(3-MERCAPTO-PROPYL)-PROPIONAMIDE, interleukin-2 | | Authors: | Arkin, M.A, Randal, M, DeLano, W.L, Hyde, J, Luong, T.N, Oslob, J.D, Raphael, D.R, Taylor, L, Wang, J, McDowell, R.S, Wells, J.A, Braisted, A.C. | | Deposit date: | 2002-07-02 | | Release date: | 2002-07-31 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Binding of small molecules to an adaptive
protein-protein interface
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1M4C
 
 | | Crystal Structure of Human Interleukin-2 | | Descriptor: | interleukin-2 | | Authors: | Arkin, M.A, Randal, M, DeLano, W.L, Hyde, J, Luong, T.N, Oslob, J.D, Raphael, D.R, Taylor, L, Wang, J, McDowell, R.S, Wells, J.A, Braisted, A.C. | | Deposit date: | 2002-07-02 | | Release date: | 2002-07-31 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Binding of small molecules to an adaptive
protein-protein interface
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1M4D
 
 | | Aminoglycoside 2'-N-acetyltransferase from Mycobacterium tuberculosis-Complex with Coenzyme A and Tobramycin | | Descriptor: | 3'-PHOSPHATE-ADENOSINE-5'-DIPHOSPHATE, Aminoglycoside 2'-N-acetyltransferase, COENZYME A, ... | | Authors: | Vetting, M.W, Hegde, S.S, Javid-Majd, F, Blanchard, J.S, Roderick, S.L. | | Deposit date: | 2002-07-02 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Aminoglycoside 2'-N-acetyltransferase from Mycobacterium tuberculosis in complex with coenzyme A and aminoglycoside substrates.
Nat.Struct.Biol., 9, 2002
|
|
1M4G
 
 | | Aminoglycoside 2'-N-acetyltransferase from Mycobacterium tuberculosis-Complex with Coenzyme A and Ribostamycin | | Descriptor: | 3'-PHOSPHATE-ADENOSINE-5'-DIPHOSPHATE, Aminoglycoside 2'-N-acetyltransferase, COENZYME A, ... | | Authors: | Vetting, M.W, Hegde, S.S, Javid-Majd, F, Blanchard, J.S, Roderick, S.L. | | Deposit date: | 2002-07-02 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Aminoglycoside 2'-N-acetyltransferase from Mycobacterium tuberculosis in complex with coenzyme A and aminoglycoside substrates.
Nat.Struct.Biol., 9, 2002
|
|
1M4H
 
 | | Crystal Structure of Beta-secretase complexed with Inhibitor OM00-3 | | Descriptor: | Inhibitor OM00-3, beta-Secretase | | Authors: | Hong, L, Turner, R.T, Koelsch, G, Ghosh, A.K, Tang, J. | | Deposit date: | 2002-07-02 | | Release date: | 2002-08-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Memapsin 2 (beta-Secretase) in Complex with Inhibitor OM00-3
Biochemistry, 41, 2002
|
|
1M4I
 
 | | Aminoglycoside 2'-N-acetyltransferase from Mycobacterium tuberculosis-Complex with Coenzyme A and Kanamycin A | | Descriptor: | 3'-PHOSPHATE-ADENOSINE-5'-DIPHOSPHATE, Aminoglycoside 2'-N-acetyltransferase, COENZYME A, ... | | Authors: | Vetting, M.W, Hegde, S.S, Javid-Majd, F, Blanchard, J.S, Roderick, S.L. | | Deposit date: | 2002-07-02 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Aminoglycoside 2'-N-acetyltransferase from Mycobacterium tuberculosis in complex with coenzyme A and aminoglycoside substrates.
Nat.Struct.Biol., 9, 2002
|
|
1M4J
 
 | | CRYSTAL STRUCTURE OF THE N-TERMINAL ADF-H DOMAIN OF MOUSE TWINFILIN ISOFORM-1 | | Descriptor: | A6 gene product | | Authors: | Paavilainen, V.O, Merckel, M.C, Falck, S, Ojala, P.J, Pohl, E, Wilmanns, M, Lappalainen, P. | | Deposit date: | 2002-07-03 | | Release date: | 2002-11-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Conservation Between the Actin Monomer-binding Sites of Twinfilin and Actin-depolymerizing Factor (ADF)/Cofilin
J.Biol.Chem., 277, 2002
|
|
1M4K
 
 | | Crystal structure of the human natural killer cell activator receptor KIR2DS2 (CD158j) | | Descriptor: | 1,2-ETHANEDIOL, KILLER CELL IMMUNOGLOBULIN-LIKE RECEPTOR 2DS2, SULFATE ION | | Authors: | Saulquin, X, Gastinel, L.N, Vivier, E. | | Deposit date: | 2002-07-03 | | Release date: | 2003-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the human natural killer cell
activating receptor KIR2DS2 (CD158j)
J.EXP.MED., 197, 2003
|
|
1M4L
 
 | | STRUCTURE OF NATIVE CARBOXYPEPTIDASE A AT 1.25 RESOLUTION | | Descriptor: | CARBOXYPEPTIDASE A, ZINC ION | | Authors: | Kilshtain-Vardi, A, Glick, M, Greenblatt, H.M, Goldblum, A, Shoham, G. | | Deposit date: | 2002-07-03 | | Release date: | 2003-01-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Refined structure of bovine carboxypeptidase A at 1.25 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1M4N
 
 | | CRYSTAL STRUCTURE OF APPLE ACC SYNTHASE IN COMPLEX WITH [2-(AMINO-OXY)ETHYL](5'-DEOXYADENOSIN-5'-YL)(METHYL)SULFONIUM | | Descriptor: | (2-AMINOOXY-ETHYL)-[5-(6-AMINO-PURIN-9-YL)-3,4-DIHYDROXY-TETRAHYDRO-FURAN-2-YLMETHYL]-METHYL-SULFONIUM, 1-aminocyclopropane-1-carboxylate synthase, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Capitani, G, Eliot, A.C, Gut, H, Khomutov, R.M, Kirsch, J.F, Grutter, M.G. | | Deposit date: | 2002-07-03 | | Release date: | 2003-04-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of 1-aminocyclopropane-1-carboxylate synthase in complex with an amino-oxy analogue of the substrate: implications for substrate binding.
BIOCHEM.BIOPHYS.ACTA PROTEINS & PROTEOMICS, 1647, 2003
|
|
1M4R
 
 | | CRYSTAL STRUCTURE OF RECOMBINANT HUMAN INTERLEUKIN-22 | | Descriptor: | Interleukin-22 | | Authors: | Nagem, R.A.P, Colau, D, Dumoutier, L, Renauld, J.-C, Ogata, C, Polikarpov, I. | | Deposit date: | 2002-07-03 | | Release date: | 2003-07-07 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Recombinant Human Interleukin-22
Structure, 10, 2002
|
|
1M4S
 
 | | Biosynthetic thiolase, Cys89 acetylated, unliganded form | | Descriptor: | Acetyl-CoA acetyltransferase, GLYCEROL, SULFATE ION | | Authors: | Kursula, P, Ojala, J, Lambeir, A.-M, Wierenga, R.K. | | Deposit date: | 2002-07-03 | | Release date: | 2002-11-29 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The catalytic cycle of biosynthetic thiolase: A conformational
journey of an acetyl group through four binding modes and two oxyanion holes
Biochemistry, 41, 2002
|
|
1M4T
 
 | | Biosynthetic thiolase, Cys89 butyrylated | | Descriptor: | Acetyl-CoA acetyltransferase, GLYCEROL, SULFATE ION | | Authors: | Kursula, P, Ojala, J, Lambeir, A.-M, Wierenga, R.K. | | Deposit date: | 2002-07-03 | | Release date: | 2002-11-29 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The catalytic cycle of biosynthetic thiolase: A conformational
journey of an acetyl group through four binding modes and two oxyanion holes
Biochemistry, 41, 2002
|
|
1M4U
 
 | | Crystal structure of Bone Morphogenetic Protein-7 (BMP-7) in complex with the secreted antagonist Noggin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Bone Morphogenetic Protein-7, Noggin | | Authors: | Groppe, J, Greenwald, J, Wiater, E, Rodriguez-Leon, J, Economides, A.N, Kwiatkowski, W, Affolter, M, Vale, W.W, Izpisua-Belmonte, J.C, Choe, S. | | Deposit date: | 2002-07-03 | | Release date: | 2002-12-18 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural Basis of BMP Signalling Inhibition by the Cystine Knot Protein Noggin
Nature, 420, 2002
|
|
1M4V
 
 | | Crystal structure of SET3, a superantigen-like protein from Staphylococcus aureus | | Descriptor: | SET3, superantigen-like protein | | Authors: | Arcus, V.L, Langley, R, Proft, T, Fraser, J.D, Baker, E.N. | | Deposit date: | 2002-07-05 | | Release date: | 2002-07-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The three-dimensional structure of a superantigen-like protein, SET3, from a pathogenicity island of the Staphylococcus aureus genome
J.Biol.Chem., 277, 2002
|
|
1M4W
 
 | | Thermophilic b-1,4-xylanase from Nonomuraea flexuosa | | Descriptor: | ACETATE ION, GLYCEROL, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hakulinen, N, Turunen, O, Janis, J, Leisola, M, Rouvinen, J. | | Deposit date: | 2002-07-05 | | Release date: | 2003-07-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three-dimensional structures of thermophilic beta-1,4-xylanases from Chaetomium thermophilum and Nonomuraea flexuosa. Comparison of twelve xylanases in relation to their thermal stability.
Eur.J.Biochem., 270, 2003
|
|
1M4Y
 
 | | Crystal structure of HslV from Thermotoga maritima | | Descriptor: | ATP-dependent protease hslV, SODIUM ION | | Authors: | Song, H.K, Ramachandran, R, Bochtler, M.B, Hartmann, C, Azim, M.K, Huber, R. | | Deposit date: | 2002-07-05 | | Release date: | 2003-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Isolation and characterization of the prokaryotic proteasome homolog HslVU (ClpQY) from Thermotoga maritima and the crystal structure of HslV.
BIOPHYS.CHEM., 100, 2003
|
|
1M4Z
 
 | | Crystal structure of the N-terminal BAH domain of Orc1p | | Descriptor: | MANGANESE (II) ION, ORIGIN RECOGNITION COMPLEX SUBUNIT 1 | | Authors: | Zhang, Z, Hayashi, M.K, Merkel, O, Stillman, B, Xu, R.-M. | | Deposit date: | 2002-07-05 | | Release date: | 2002-09-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function of the BAH-containing domain of Orc1p in epigenetic silencing.
EMBO J., 21, 2002
|
|
1M52
 
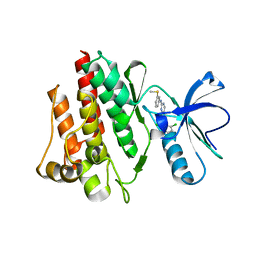 | | Crystal Structure of the c-Abl Kinase domain in complex with PD173955 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-(2,6-DICHLORO-PHENYL)-8-METHYL-2-(3-METHYLSULFANYL-PHENYLAMINO)-8H-PYRIDO[2,3-D]PYRIMIDIN-7-ONE, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE ABL1 | | Authors: | Nagar, B, Bornmann, W, Pellicena, P, Schindler, T, Veach, D, Miller, W.T, Clarkson, B, Kuriyan, J. | | Deposit date: | 2002-07-08 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of the Kinase Domain of c-Abl in Complex with the Small Molecule Inhibitors PD173955 and Imatinib (STI-571)
Cancer Res., 62, 2002
|
|
1M53
 
 | |
1M54
 
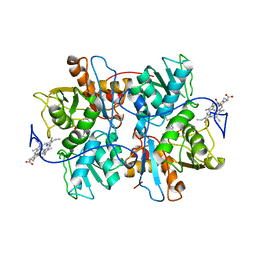 | | CYSTATHIONINE-BETA SYNTHASE: REDUCED VICINAL THIOLS | | Descriptor: | CYSTATHIONINE BETA-SYNTHASE, PROTOPORPHYRIN IX CONTAINING FE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Taoka, S, Lepore, B.W, Kabil, O, Ojha, S, Ringe, D, Banerjee, R. | | Deposit date: | 2002-07-08 | | Release date: | 2002-08-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | HUMAN CYSTATHIONINE BETA-SYNTHASE IS A HEME SENSOR PROTEIN. EVIDENCE THAT THE
REDOX SENSOR IS HEME AND NOT THE VICINAL CYSTEINES IN THE CXXC MOTIF SEEN IN THE CRYSTAL STRUCTURE OF THE TRUNCATED ENZYME
BIOCHEMISTRY, 41, 2002
|
|
1M59
 
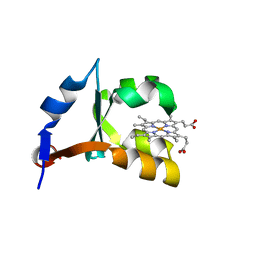 | | Crystal Structure of P40V Mutant of Trypsin-solubilized Fragment of Cytochrome b5 | | Descriptor: | Cytochrome b5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wang, Z.-Q, Wu, J, Wang, Y.-H, Qian, W, Xie, Y, Xia, Z.-X, Huang, Z.-X. | | Deposit date: | 2002-07-09 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Proline40 is essential to maintaining cytochrome b5 stability and its electron transfer with cytochrome c
Chin.J.Chem., 20, 2002
|
|
1M5B
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH 2-Me-Tet-AMPA AT 1.85 A RESOLUTION. | | Descriptor: | (S)-2-AMINO-3-[3-HYDROXY-5-(2-METHYL-2H-TETRAZOL-5-YL)ISOXAZOL-4-YL]PROPIONIC ACID, Glutamate receptor 2, ZINC ION | | Authors: | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | Deposit date: | 2002-07-09 | | Release date: | 2002-09-18 | | Last modified: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
1M5C
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH Br-HIBO AT 1.65 A RESOLUTION | | Descriptor: | (S)-2-AMINO-3-(4-BROMO-3-HYDROXY-ISOXAZOL-5-YL)PROPIONIC ACID, Glutamate receptor 2 | | Authors: | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | Deposit date: | 2002-07-09 | | Release date: | 2002-09-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
