1K8C
 
 | | Crystal structure of dimeric xylose reductase in complex with NADP(H) | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, xylose reductase | | Authors: | Kavanagh, K.L, Klimacek, M, Nidetzky, B, Wilson, D.K. | | Deposit date: | 2001-10-23 | | Release date: | 2002-07-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of apo and holo forms of xylose reductase, a dimeric aldo-keto reductase from Candida tenuis.
Biochemistry, 41, 2002
|
|
1K8F
 
 | | CRYSTAL STRUCTURE OF THE HUMAN C-TERMINAL CAP1-ADENYLYL CYCLASE ASSOCIATED PROTEIN | | Descriptor: | ADENYLYL CYCLASE-ASSOCIATED PROTEIN | | Authors: | Patskovsky, Y.V, Chance, M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2001-10-24 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the actin binding domain of the cyclase-associated protein.
Biochemistry, 43, 2004
|
|
1K8G
 
 | |
1K8K
 
 | | Crystal Structure of Arp2/3 Complex | | Descriptor: | ACTIN-LIKE PROTEIN 2, ACTIN-LIKE PROTEIN 3, ARP2/3 COMPLEX 16 KDA SUBUNIT, ... | | Authors: | Robinson, R.C, Turbedsky, K, Kaiser, D.A, Higgs, H.N, Marchand, J.-B, Choe, S, Pollard, T.D. | | Deposit date: | 2001-10-24 | | Release date: | 2001-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Arp2/3 Complex
Science, 294, 2001
|
|
1K8P
 
 | |
1K8Q
 
 | | CRYSTAL STRUCTURE OF DOG GASTRIC LIPASE IN COMPLEX WITH A PHOSPHONATE INHIBITOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Triacylglycerol lipase, gastric, ... | | Authors: | Roussel, A, Miled, N, Berti-Dupuis, L, Riviere, M, Spinelli, S, Berna, P, Gruber, V, Verger, R, Cambillau, C. | | Deposit date: | 2001-10-25 | | Release date: | 2002-03-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the open form of dog gastric lipase in complex with a phosphonate inhibitor.
J.Biol.Chem., 277, 2002
|
|
1K8R
 
 | | Crystal structure of Ras-Bry2RBD complex | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Protein kinase byr2, ... | | Authors: | Scheffzek, K, Gruenewald, P, Wohlgemuth, S, Kabsch, W, Tu, H, Wigler, M, Wittinghofer, A, Herrmann, C. | | Deposit date: | 2001-10-25 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Ras-Byr2RBD complex: structural basis for Ras effector recognition in yeast.
Structure, 9, 2001
|
|
1K8T
 
 | | Crystal structure of the adenylyl cyclase domain of anthrax edema factor (EF) | | Descriptor: | CALMODULIN-SENSITIVE ADENYLATE CYCLASE, NICKEL (II) ION, SULFATE ION | | Authors: | Drum, C.L, Yan, S.-Z, Bard, J, Shen, Y.-Q, Lu, D, Soelaiman, S, Grabarek, Z, Bohm, A, Tang, W.-J. | | Deposit date: | 2001-10-25 | | Release date: | 2002-01-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the activation of anthrax adenylyl cyclase exotoxin by calmodulin
Nature, 415, 2002
|
|
1K8W
 
 | |
1K8X
 
 | | Crystal Structure Of AlphaT183V Mutant Of Tryptophan Synthase From Salmonella Typhimurium | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, TRYPTOPHAN SYNTHASE, ... | | Authors: | Kulik, V, Weyand, M, Siedel, R, Niks, D, Arac, D, Dunn, M.F, Schlichting, I. | | Deposit date: | 2001-10-26 | | Release date: | 2002-12-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | On the Role of AlphaThr183 in the Allosteric Regulation and Catalytic Mechanism of Tryptophan Synthase
J.Mol.Biol., 324, 2002
|
|
1K8Y
 
 | | CRYSTAL STRUCTURE OF THE TRYPTOPHAN SYNTHASE BETA-SER178PRO MUTANT COMPLEXED WITH D,L-ALPHA-GLYCEROL-3-PHOSPHATE | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Weyand, M, Schlichting, I, Marabotti, A, Mozzarelli, A. | | Deposit date: | 2001-10-26 | | Release date: | 2002-06-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the beta Ser178--> Pro mutant of tryptophan synthase. A "knock-out" allosteric enzyme.
J.Biol.Chem., 277, 2002
|
|
1K8Z
 
 | | CRYSTAL STRUCTURE OF THE TRYPTOPHAN SYNTHASE BETA-SER178PRO MUTANT COMPLEXED WITH N-[1H-INDOL-3-YL-ACETYL]GLYCINE ACID | | Descriptor: | N-[1H-INDOL-3-YL-ACETYL]GLYCINE ACID, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Weyand, M, Schlichting, I, Marabotti, A, Mozzarelli, A. | | Deposit date: | 2001-10-26 | | Release date: | 2002-06-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the beta Ser178--> Pro mutant of tryptophan synthase. A "knock-out" allosteric enzyme.
J.Biol.Chem., 277, 2002
|
|
1K90
 
 | | Crystal structure of the adenylyl cyclase domain of anthrax edema factor (EF) in complex with calmodulin and 3' deoxy-ATP | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, CALMODULIN, ... | | Authors: | Drum, C.L, Yan, S.-Z, Bard, J, Shen, Y.-Q, Lu, D, Soelaiman, S, Grabarek, Z, Bohm, A, Tang, W.-J. | | Deposit date: | 2001-10-26 | | Release date: | 2002-01-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for the activation of anthrax adenylyl cyclase exotoxin by calmodulin.
Nature, 415, 2002
|
|
1K92
 
 | |
1K93
 
 | | Crystal structure of the adenylyl cyclase domain of anthrax edema factor (EF) in complex with calmodulin | | Descriptor: | CALCIUM ION, CALMODULIN, CALMODULIN-SENSITIVE ADENYLATE CYCLASE, ... | | Authors: | Drum, C.L, Yan, S.-Z, Bard, J, Shen, Y.-Q, Lu, D, Soelaiman, S, Grabarek, Z, Bohm, A, Tang, W.-J. | | Deposit date: | 2001-10-26 | | Release date: | 2002-01-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis for the activation of anthrax adenylyl cyclase exotoxin by calmodulin.
Nature, 415, 2002
|
|
1K94
 
 | | Crystal structure of des(1-52)grancalcin with bound calcium | | Descriptor: | CALCIUM ION, GRANCALCIN | | Authors: | Jia, J, Borregaard, N, Lollike, K, Cygler, M. | | Deposit date: | 2001-10-26 | | Release date: | 2001-12-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Ca(2+)-loaded human grancalcin.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1K95
 
 | | Crystal structure of des(1-52)grancalcin with bound calcium | | Descriptor: | GRANCALCIN | | Authors: | Jia, J, Borregaard, N, Lollike, K, Cygler, M. | | Deposit date: | 2001-10-26 | | Release date: | 2001-12-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Ca(2+)-loaded human grancalcin.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1K97
 
 | |
1K9A
 
 | | Crystal structure analysis of full-length carboxyl-terminal Src kinase at 2.5 A resolution | | Descriptor: | Carboxyl-terminal Src kinase | | Authors: | Ogawa, A, Takayama, Y, Nagata, A, Chong, K.T, Takeuchi, S, Sakai, H, Nakagawa, A, Nada, S, Okada, M, Tsukihara, T. | | Deposit date: | 2001-10-28 | | Release date: | 2002-03-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the carboxyl-terminal Src kinase, Csk.
J.Biol.Chem., 277, 2002
|
|
1K9I
 
 | | Complex of DC-SIGN and GlcNAc2Man3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose, CALCIUM ION, mDC-SIGN1B type I isoform | | Authors: | Feinberg, H, Mitchell, D.A, Drickamer, K, Weis, W.I. | | Deposit date: | 2001-10-29 | | Release date: | 2001-12-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for selective recognition of oligosaccharides by DC-SIGN and DC-SIGNR.
Science, 294, 2001
|
|
1K9J
 
 | | Complex of DC-SIGNR and GlcNAc2Man3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose, CALCIUM ION, mDC-SIGN2 TYPE I ISOFORM | | Authors: | Feinberg, H, Mitchell, D.A, Drickamer, K, Weis, W.I. | | Deposit date: | 2001-10-29 | | Release date: | 2001-12-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for selective recognition of oligosaccharides by DC-SIGN and DC-SIGNR.
Science, 294, 2001
|
|
1K9M
 
 | | Co-crystal structure of tylosin bound to the 50S ribosomal subunit of Haloarcula marismortui | | Descriptor: | 23S RRNA, 5S RRNA, CADMIUM ION, ... | | Authors: | Hansen, J.L, Ippolito, J.A, Ban, N, Nissen, P, Moore, P.B, Steitz, T.A. | | Deposit date: | 2001-10-29 | | Release date: | 2002-07-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structures of four macrolide antibiotics bound to the large ribosomal subunit.
Mol.Cell, 10, 2002
|
|
1K9O
 
 | | CRYSTAL STRUCTURE OF MICHAELIS SERPIN-TRYPSIN COMPLEX | | Descriptor: | ALASERPIN, TRYPSIN II ANIONIC | | Authors: | Ye, S, Cech, A.L, Belmares, R, Bergstrom, R.C, Tong, Y, Corey, D.R, Kanost, M.R, Goldsmith, E.J. | | Deposit date: | 2001-10-29 | | Release date: | 2001-11-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of a Michaelis serpin-protease complex.
Nat.Struct.Biol., 8, 2001
|
|
1K9T
 
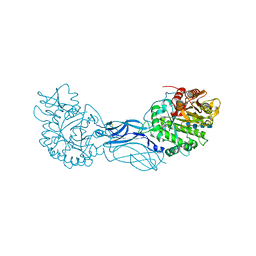 | | Chitinase a complexed with tetra-N-acetylchitotriose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITINASE A | | Authors: | Prag, G, Tucker, P.A, Oppenheim, A.B. | | Deposit date: | 2001-10-30 | | Release date: | 2002-11-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Complex Structures of Chitinase A Mutant with Oligonag Provide Insight Into the Enzymatic Mechanism
To be Published
|
|
1K9U
 
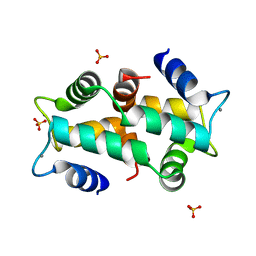 | | Crystal Structure of the Calcium-Binding Pollen Allergen Phl p 7 (Polcalcin) at 1.75 Angstroem | | Descriptor: | CALCIUM ION, Polcalcin Phl p 7, SULFATE ION | | Authors: | Verdino, P, Westritschnig, K, Valenta, R, Keller, W. | | Deposit date: | 2001-10-30 | | Release date: | 2003-04-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The cross-reactive calcium-binding pollen allergen, Phl p 7, reveals a novel dimer assembly
EMBO J., 21, 2002
|
|
