1PXM
 
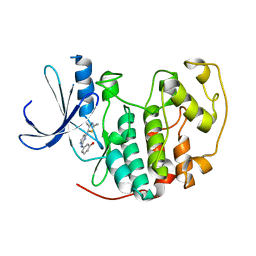 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE INHIBITOR 3-[4-(2,4-Dimethyl-thiazol-5-yl)-pyrimidin-2-ylamino]-phenol | | Descriptor: | 3-[4-(2,4-DIMETHYL-THIAZOL-5-YL)-PYRIMIDIN-2-YLAMINO]-PHENOL, Cell division protein kinase 2 | | Authors: | Wang, S, Meades, C, Wood, G, Osnowski, A, Anderson, S, Yuill, R, Thomas, M, Jackson, W, Midgley, C, Griffiths, G, McNae, I, Wu, S.Y, McInnes, C, Zheleva, D, Walkinshaw, M.D, Fischer, P.M. | | Deposit date: | 2003-07-04 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | 2-Anilino-4-(thiazol-5-yl)pyrimidine CDK inhibitors: synthesis, SAR analysis, X-ray crystallography, and biological activity.
J.Med.Chem., 47, 2004
|
|
1PXN
 
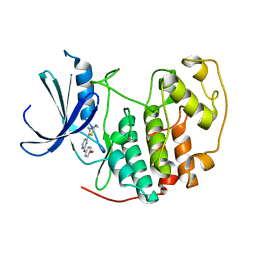 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE INHIBITOR 4-[4-(4-Methyl-2-methylamino-thiazol-5-yl)-pyrimidin-2-ylamino]-phenol | | Descriptor: | 4-[4-(4-METHYL-2-METHYLAMINO-THIAZOL-5-YL)-PYRIMIDIN-2-YLAMINO]-PHENOL, Cell division protein kinase 2 | | Authors: | Wang, S, Meades, C, Wood, G, Osnowski, A, Anderson, S, Yuill, R, Thomas, M, Mezna, M, Jackson, W, Midgley, C, Griffiths, G, McNae, I, Wu, S.Y, McInnes, C, Zheleva, D, Walkinshaw, M.D, Fischer, P.M. | | Deposit date: | 2003-07-04 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 2-Anilino-4-(thiazol-5-yl)pyrimidine CDK inhibitors: synthesis, SAR analysis, X-ray crystallography, and biological activity.
J.Med.Chem., 47, 2004
|
|
1PXO
 
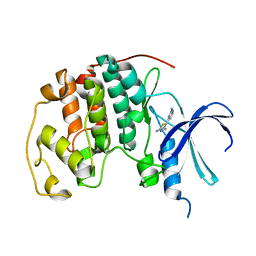 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE INHIBITOR [4-(2-Amino-4-methyl-thiazol-5-yl)-pyrimidin-2-yl]-(3-nitro-phenyl)-amine | | Descriptor: | Cell division protein kinase 2, [4-(2-AMINO-4-METHYL-THIAZOL-5-YL)-PYRIMIDIN-2-YL]-(3-NITRO-PHENYL)-AMINE | | Authors: | Wang, S, Meades, C, Wood, G, Osnowski, A, Anderson, S, Yuill, R, Thomas, M, Mezna, M, Jackson, W, Midgley, C, Griffiths, G, McNae, I, Wu, S.Y, McInnes, C, Zheleva, D, Walkinshaw, M.D, Fischer, P.M. | | Deposit date: | 2003-07-04 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | 2-Anilino-4-(thiazol-5-yl)pyrimidine CDK inhibitors: synthesis, SAR analysis, X-ray crystallography, and biological activity.
J.Med.Chem., 47, 2004
|
|
1PXP
 
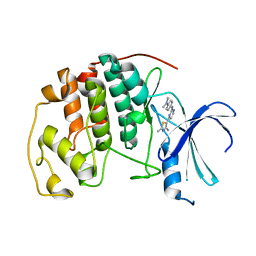 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE INHIBITOR N-[4-(2,4-Dimethyl-thiazol-5-yl)-pyrimidin-2-yl]-N',N'-dimethyl-benzene-1,4-diamine | | Descriptor: | Cell division protein kinase 2, N-[4-(2,4-DIMETHYL-THIAZOL-5-YL)-PYRIMIDIN-2-YL]-N',N'-DIMETHYL-BENZENE-1,4-DIAMINE | | Authors: | Wang, S, Meades, C, Wood, G, Osnowski, A, Anderson, S, Yuill, R, Thomas, M, Mezna, M, Jackson, W, Midgley, C, Griffiths, G, McNae, I, Wu, S.Y, McInnes, C, Zheleva, D, Walkinshaw, M.D, Fischer, P.M. | | Deposit date: | 2003-07-04 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 2-Anilino-4-(thiazol-5-yl)pyrimidine CDK inhibitors: synthesis, SAR analysis, X-ray crystallography, and biological activity.
J.Med.Chem., 47, 2004
|
|
1PXT
 
 | |
1PXU
 
 | |
1PXV
 
 | | The staphostatin-staphopain complex: a forward binding inhibitor in complex with its target cysteine protease | | Descriptor: | GUANIDINE, SULFATE ION, cysteine protease, ... | | Authors: | Filipek, R, Rzychon, M, Oleksy, A, Gruca, M, Dubin, A, Potempa, J, Bochtler, M. | | Deposit date: | 2003-07-07 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Staphostatin-Staphopain Complex: A FORWARD BINDING INHIBITOR IN COMPLEX WITH ITS TARGET CYSTEINE PROTEASE.
J.Biol.Chem., 278, 2003
|
|
1PXW
 
 | | Crystal structure of L7Ae sRNP core protein from Pyrococcus abyssii | | Descriptor: | LSU ribosomal protein L7AE | | Authors: | Charron, C, Manival, X, Charpentier, B, Branlant, C, Aubry, A. | | Deposit date: | 2003-07-07 | | Release date: | 2004-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Purification, crystallization and preliminary X-ray diffraction data of L7Ae sRNP core protein from Pyrococcus abyssii.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1PXX
 
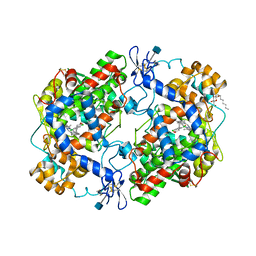 | | CRYSTAL STRUCTURE OF DICLOFENAC BOUND TO THE CYCLOOXYGENASE ACTIVE SITE OF COX-2 | | Descriptor: | 2-[2,6-DICHLOROPHENYL)AMINO]BENZENEACETIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kiefer, J.R, Rowlinson, S.W, Prusakiewicz, J.J, Pawlitz, J.L, Kozak, K.R, Kalgutkar, A.S, Stallings, W.C, Marnett, L.J, Kurumbail, R.G. | | Deposit date: | 2003-07-07 | | Release date: | 2003-09-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A Novel Mechanism of Cyclooxygenase-2 Inhibition Involving Interactions with Ser-530 and Tyr-385.
J.Biol.Chem., 278, 2003
|
|
1PXY
 
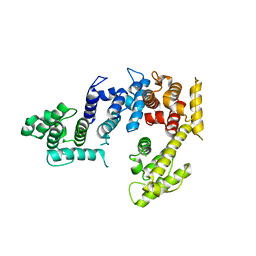 | | Crystal structure of the actin-crosslinking core of Arabidopsis fimbrin | | Descriptor: | fimbrin-like protein | | Authors: | Klein, M.G, Shi, W, Tseng, Y, Wirtz, D, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-07-07 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the actin crosslinking core of fimbrin.
Structure, 12, 2004
|
|
1PXZ
 
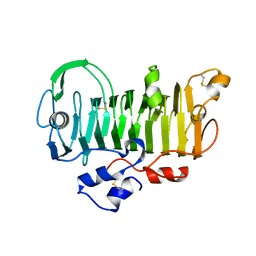 | | 1.7 Angstrom Crystal Structure of jun a 1, the major allergen from cedar pollen | | Descriptor: | Major pollen allergen Jun a 1 | | Authors: | Czerwinski, E.W, White, M.A, Midoro-Horiuti, T, Brooks, E.G, Goldblum, R.M. | | Deposit date: | 2003-07-07 | | Release date: | 2004-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Jun a 1, the major cedar pollen allergen from Juniperus ashei, reveals a parallel beta-helical core.
J.Biol.Chem., 280, 2005
|
|
1PY1
 
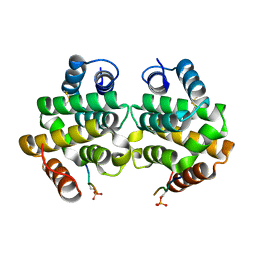 | | Complex of GGA1-VHS domain and beta-secretase C-terminal phosphopeptide | | Descriptor: | ADP-ribosylation factor binding protein GGA1, Beta-secretase | | Authors: | Zhu, G, Zhang, X.C. | | Deposit date: | 2003-07-07 | | Release date: | 2003-11-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Biochemical and structural characterization of the interaction of memapsin 2 (beta-secretase)
cytosolic domain with the VHS domain of GGA proteins.
Biochemistry, 42, 2003
|
|
1PY2
 
 | | Structure of a 60 nM Small Molecule Bound to a Hot Spot on IL-2 | | Descriptor: | 5-[2,3-DICHLORO-4-(5-{1-[2-(2-GUANIDINO-4-METHYL-PENTANOYLAMINO)-ACETYL]-PIPERIDIN-4-YL}-1-METHYL-1H-PYRAZOL-3-YL)-PHENOXYMETHYL]-FURAN-2-CARBOXYLIC ACID, Interleukin-2, ZINC ION | | Authors: | Thanos, C.D, Randal, M, Wells, J.A. | | Deposit date: | 2003-07-07 | | Release date: | 2004-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Potent small-molecule binding to a dynamic hot spot on IL-2.
J.Am.Chem.Soc., 125, 2003
|
|
1PY3
 
 | | Crystal structure of Ribonuclease Sa2 | | Descriptor: | SULFATE ION, ribonuclease | | Authors: | Sevcik, J, Dauter, Z, Wilson, K.S. | | Deposit date: | 2003-07-08 | | Release date: | 2004-07-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure reveals two alternative conformations in the active site of ribonuclease Sa2.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1PY4
 
 | | Beta2 microglobulin mutant H31Y displays hints for amyloid formations | | Descriptor: | Beta-2-microglobulin precursor | | Authors: | Rosano, C, Zuccotti, S, Mangione, P, Giorgetti, S, Bellotti, V, Pettirossi, F, Corazza, A, Viglino, P, Esposito, G, Bolognesi, M. | | Deposit date: | 2003-07-08 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | beta2-microglobulin H31Y variant 3D structure highlights the protein natural propensity towards intermolecular aggregation
J.Mol.Biol., 335, 2004
|
|
1PY9
 
 | | The crystal structure of an autoantigen in multiple sclerosis | | Descriptor: | Myelin-oligodendrocyte glycoprotein, SULFATE ION | | Authors: | Clements, C.S, Reid, H.H, Beddoe, T, Tynan, F.E, Perugini, M.A, Johns, T.G, Bernard, C.C, Rossjohn, J. | | Deposit date: | 2003-07-08 | | Release date: | 2003-09-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of myelin oligodendrocyte glycoprotein, a key autoantigen in multiple sclerosis
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1PYA
 
 | |
1PYB
 
 | | Crystal Structure of Aquifex aeolicus Trbp111: a Structure-Specific tRNA Binding Protein | | Descriptor: | tRNA-binding protein Trbp111 | | Authors: | Swairjo, M.A, Morales, A.J, Wang, C.C, Ortiz, A.R, Schimmel, P. | | Deposit date: | 2003-07-08 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of trbp111: a structure-specific tRNA-binding protein.
Embo J., 19, 2000
|
|
1PYF
 
 | |
1PYH
 
 | | Crystal structure of RC-LH1 core complex from Rhodopseudomonas palustris | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Roszak, A.W, Howard, T.D, Southall, J, Gardiner, A.T, Law, C.J, Isaacs, N.W, Cogdell, R.J. | | Deposit date: | 2003-07-08 | | Release date: | 2004-03-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (4.8 Å) | | Cite: | Crystal structure of the RC-LH1 core complex from Rhodopseudomonas palustris.
Science, 302, 2003
|
|
1PYI
 
 | |
1PYL
 
 | | Crystal structure of Ribonuclease Sa2 | | Descriptor: | SULFATE ION, ribonuclease | | Authors: | Sevcik, J, Dauter, Z, Wilson, K.S. | | Deposit date: | 2003-07-09 | | Release date: | 2004-07-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.507 Å) | | Cite: | Crystal structure reveals two alternative conformations in the active site of ribonuclease Sa2.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1PYO
 
 | |
1PYQ
 
 | | Unprocessed Aspartate Decarboxylase Mutant, with Alanine inserted at position 24 | | Descriptor: | Aspartate 1-decarboxylase, SULFATE ION | | Authors: | Schmitzberger, F, Kilkenny, M.L, Lobley, C.M.C, Webb, M.E, Vinkovic, M, Matak-Vinkovic, D, Witty, M, Chirgadze, D.Y, Smith, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2003-07-09 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Constraints on protein self-processing in L-aspartate-alpha-decarboxylase
Embo J., 22, 2003
|
|
1PYS
 
 | | PHENYLALANYL-TRNA SYNTHETASE FROM THERMUS THERMOPHILUS | | Descriptor: | MAGNESIUM ION, PHENYLALANYL-TRNA SYNTHETASE | | Authors: | Safro, M, Mosyak, L, Goldgur, Y, Reshetnikova, L, Delarue, M. | | Deposit date: | 1996-11-14 | | Release date: | 1997-11-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of phenylalanyl-tRNA synthetase from Thermus thermophilus.
Nat.Struct.Biol., 2, 1995
|
|
