2NDL
 
 | | NMR solution structure of PawS Derived Peptide 22 (PDP-22) | | Descriptor: | PawS derived peptide | | Authors: | Franke, B, Jayasena, A.S, Fisher, M.F, Swedberg, J.E, Taylor, N.L, Mylne, J.S, Rosengren, K. | | Deposit date: | 2016-07-17 | | Release date: | 2016-12-07 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Diverse cyclic seed peptides in the Mexican zinnia (Zinnia haageana).
Biopolymers, 106, 2016
|
|
2NDM
 
 | | NMR solution structure of PawS Derived Peptide 21 (PDP-21) | | Descriptor: | PawS derived peptide 21 | | Authors: | Franke, B, Jayasena, A.S, Fisher, M.F, Swedberg, J.E, Taylor, N.L, Mylne, J.S, Rosengren, K. | | Deposit date: | 2016-07-17 | | Release date: | 2016-08-24 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Diverse cyclic seed peptides in the Mexican zinnia (Zinnia haageana).
Biopolymers, 106, 2016
|
|
2NDN
 
 | | NMR solution structure of PawS Derived Peptide 20 (PDP-20) | | Descriptor: | PawS1a Derived Peptide 20 | | Authors: | Franke, B, Jayasena, A.S, Fisher, M.F, Swedberg, J.E, Taylor, N.L, Mylne, J.S, Rosengren, K. | | Deposit date: | 2016-07-26 | | Release date: | 2016-08-24 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Diverse cyclic seed peptides in the Mexican zinnia (Zinnia haageana).
Biopolymers, 106, 2016
|
|
2NDO
 
 | | Structure of EcDsbA-sulfonamide1 complex | | Descriptor: | 2-{[(4-iodophenyl)sulfonyl]amino}benzoic acid, Thiol:disulfide interchange protein DsbA | | Authors: | Williams, M.L, Doak, B.C, Vazirani, M, Ilyichova, O, Wang, G, Bermel, W, Simpson, J.S, Chalmers, D.K, King, G.F, Mobli, M, Scanlon, M.J. | | Deposit date: | 2016-08-22 | | Release date: | 2017-02-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Determination of ligand binding modes in weak protein-ligand complexes using sparse NMR data.
J.Biomol.Nmr, 66, 2016
|
|
2NDP
 
 | | Structure of DNA-binding HU protein from micoplasma Mycoplasma gallisepticum | | Descriptor: | Histone-like DNA-binding superfamily protein | | Authors: | Altukhov, D.A, Talyzina, A.A, Agapova, Y.K, Vlaskina, A.V, Korzhenevskiy, D.A, Bocharov, E.V, Rakitina, T.V, Timofeev, V.I, Popov, V.O. | | Deposit date: | 2016-09-13 | | Release date: | 2016-11-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Enhanced conformational flexibility of the histone-like (HU) protein from Mycoplasma gallisepticum.
J.Biomol.Struct.Dyn., 36, 2018
|
|
2NNZ
 
 | | Solution structure of the hypothetical protein AF2241 from Archaeoglobus fulgidus | | Descriptor: | Hypothetical protein | | Authors: | Ai, X, Semesib, A, Yee, A, Arrowsmith, C.H, Li, S.S.C, Choy, W.Y, Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2006-10-24 | | Release date: | 2007-08-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Hypothetical protein AF2241 from Archaeoglobus fulgidus adopts a cyclophilin-like fold.
J.Biomol.Nmr, 38, 2007
|
|
2NPB
 
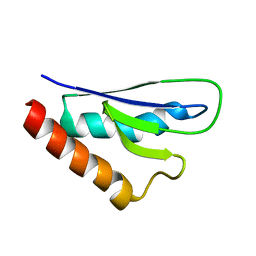 | | NMR solution structure of mouse SelW | | Descriptor: | Selenoprotein W | | Authors: | Aachmann, F.L, Fomenko, D.E, Soragni, A, Gladyshev, V.N, Dikiy, A. | | Deposit date: | 2006-10-27 | | Release date: | 2006-11-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of selenoprotein W and NMR analysis of its interaction with 14-3-3 proteins
J.Biol.Chem., 282, 2007
|
|
2NPU
 
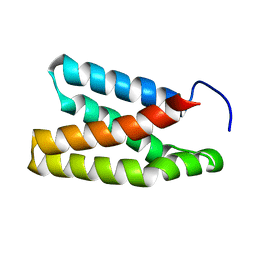 | | The solution structure of the rapamycin-binding domain of mTOR (FRB) | | Descriptor: | FKBP12-rapamycin complex-associated protein | | Authors: | Veverka, V, Crabbe, T, Bird, I, Lennie, G, Muskett, F.W, Taylor, R.J, Carr, M.D. | | Deposit date: | 2006-10-30 | | Release date: | 2007-09-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of the interaction of mTOR with phosphatidic acid and a novel class of inhibitor: compelling evidence for a central role of the FRB domain in small molecule-mediated regulation of mTOR.
Oncogene, 27, 2008
|
|
2NSV
 
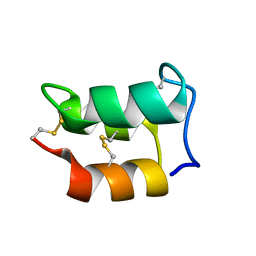 | |
2NSW
 
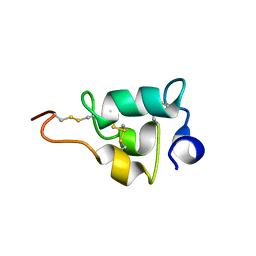 | |
2O4E
 
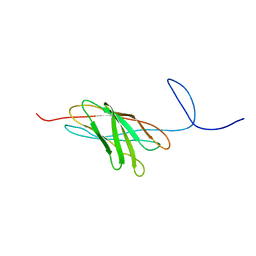 | | The solution structure of a protein-protein interaction module from a family 84 glycoside hydrolase of Clostridium perfringens | | Descriptor: | O-GlcNAcase nagJ | | Authors: | Chitayat, S, Adams, J.J, Gregg, K, Boraston, A.B, Smith, S.P. | | Deposit date: | 2006-12-04 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of a putative non-cellulosomal cohesin module from a Clostridium perfringens family 84 glycoside hydrolase.
J.Mol.Biol., 375, 2008
|
|
2OFN
 
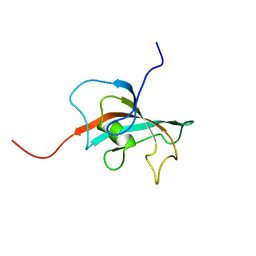 | | Solution structure of FK506-binding domain (FKBD)of FKBP35 from Plasmodium falciparum | | Descriptor: | 70 kDa peptidylprolyl isomerase, putative | | Authors: | Kang, C.B, Ye, H, Yoon, H.R, Yoon, H.S. | | Deposit date: | 2007-01-04 | | Release date: | 2007-12-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of FK506 binding domain (FKBD) of Plasmodium falciparum FK506 binding protein 35 (PfFKBP35).
Proteins, 70, 2007
|
|
2OI3
 
 | | NMR Structure Analysis of the Hematopoetic Cell Kinase SH3 Domain complexed with an artificial high affinity ligand (PD1) | | Descriptor: | Tyrosine-protein kinase HCK, artificial peptide PD1 | | Authors: | Schmidt, H, Stoldt, M, Hoffmann, S, Tran, T, Willbold, D. | | Deposit date: | 2007-01-10 | | Release date: | 2007-02-20 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Hck SH3 Domain Ligand
Complex Reveals Novel Interaction Modes
J.Mol.Biol., 365, 2007
|
|
2OV6
 
 | |
2POA
 
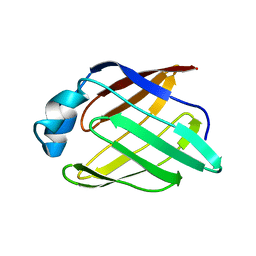 | | Schistosoma mansoni Sm14 Fatty Acid-Binding Protein: improvement of protein stability by substitution of the single Cys62 residue | | Descriptor: | 14 kDa fatty acid-binding protein | | Authors: | Ramos, C.R.R, Oyama Jr, S, Sforca, M.L, Pertinhez, T.A, Ho, P.L, Spisni, A. | | Deposit date: | 2007-04-26 | | Release date: | 2008-06-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Stability improvement of the fatty acid binding protein Sm14 from S. mansoni by Cys replacement: Structural and functional characterization of a vaccine candidate.
Biochim.Biophys.Acta, 1794, 2009
|
|
2PXG
 
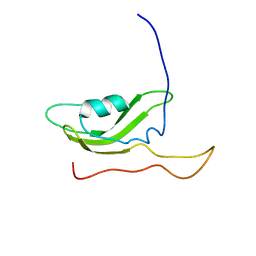 | | NMR Solution Structure of OmlA | | Descriptor: | Outer membrane protein | | Authors: | Vanini, M.M.T, Pertinhez, T.A, Sforca, M.L, Spisni, A, Benedetti, C.E. | | Deposit date: | 2007-05-14 | | Release date: | 2008-01-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the outer membrane lipoprotein OmlA from Xanthomonas axonopodis pv. citri reveals a protein fold implicated in protein-protein interaction.
Proteins, 71, 2008
|
|
2RLY
 
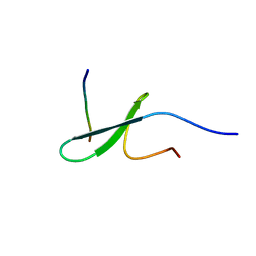 | | FBP28WW2 domain in complex with PTPPPLPP peptide | | Descriptor: | Formin-1, Transcription elongation regulator 1 | | Authors: | Ramirez-Espain, X, Ruiz, L, Martin-Malpartida, P, Oschkinat, H, Macias, M.J. | | Deposit date: | 2007-09-03 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of a New Binding Motif and a Novel Binding Mode in Group 2 WW Domains
J.Mol.Biol., 373, 2007
|
|
2RM0
 
 | | FBP28WW2 domain in complex with a PPPLIPPPP peptide | | Descriptor: | Formin-1, Transcription elongation regulator 1 | | Authors: | Ramirez-Espain, X, Ruiz, L, Martin-Malpartida, P, Oschkinat, H, Macias, M.J. | | Deposit date: | 2007-09-06 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of a New Binding Motif and a Novel Binding Mode in Group 2 WW Domains
J.Mol.Biol., 373, 2007
|
|
2RN1
 
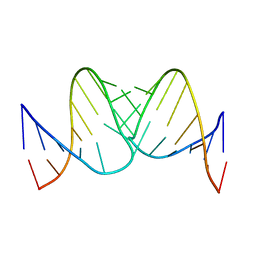 | | Liquid crystal solution structure of the kissing complex formed by the apical loop of the HIV TAR RNA and a high affinity RNA aptamer optimized by SELEX | | Descriptor: | RNA (5'-R(P*GP*AP*GP*CP*CP*CP*UP*GP*GP*GP*AP*GP*GP*CP*UP*C)-3'), RNA (5'-R(P*GP*CP*UP*GP*GP*UP*CP*CP*CP*AP*GP*AP*CP*AP*GP*C)-3') | | Authors: | Van Melckebeke, H, Devany, M, Di Primo, C, Beaurain, F, Toulme, J, Bryce, D.L, Boisbouvier, J. | | Deposit date: | 2007-12-05 | | Release date: | 2008-09-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Liquid-crystal NMR structure of HIV TAR RNA bound to its SELEX RNA aptamer reveals the origins of the high stability of the complex
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2RN8
 
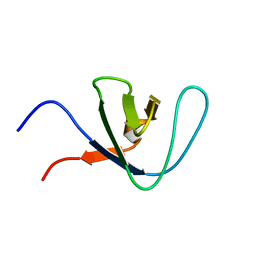 | |
2RN9
 
 | | Solution structure of human apoCox17 | | Descriptor: | Cytochrome c oxidase copper chaperone | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Janicka, A, Martinelli, M, Kozlowski, H, Palumaa, P. | | Deposit date: | 2007-12-08 | | Release date: | 2007-12-18 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | A structural-dynamical characterization of human cox17
J.Biol.Chem., 283, 2008
|
|
2RNB
 
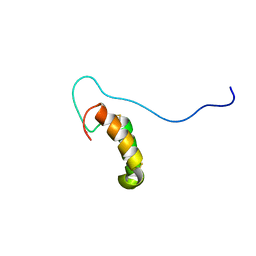 | | Solution structure of human Cu(I)Cox17 | | Descriptor: | COPPER (I) ION, Cytochrome c oxidase copper chaperone | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Janicka, A, Martinelli, M, Kozlowski, H, Palumaa, P. | | Deposit date: | 2007-12-08 | | Release date: | 2007-12-18 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | A structural-dynamical characterization of human cox17
J.Biol.Chem., 283, 2008
|
|
2RNG
 
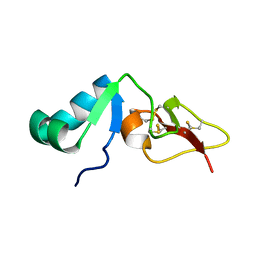 | | Solution structure of big defensin | | Descriptor: | Big defensin | | Authors: | Kouno, T, Fujitani, N, Osaki, T, Kawabata, S, Nishimura, S, Mizuguchi, M, Aizawa, T, Demura, M, Nitta, K, Kawano, K. | | Deposit date: | 2007-12-27 | | Release date: | 2008-10-14 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | A novel beta-defensin structure: a potential strategy of big defensin for overcoming resistance by Gram-positive bacteria
Biochemistry, 47, 2008
|
|
2RNN
 
 | |
2RNO
 
 | |
