2LEE
 
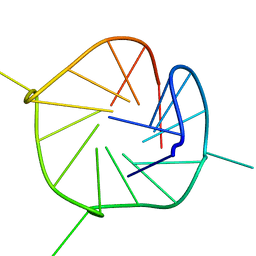 | |
2LEG
 
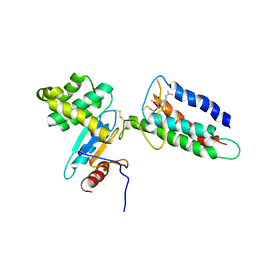 | | Membrane protein complex DsbB-DsbA structure by joint calculations with solid-state NMR and X-ray experimental data | | Descriptor: | Disulfide bond formation protein B, Thiol:disulfide interchange protein DsbA, UBIQUINONE-1, ... | | Authors: | Tang, M, Sperling, L.J, Berthold, D.A, Schwieters, C.D, Nesbitt, A.E, Nieuwkoop, A.J, Gennis, R.B, Rienstra, C.M. | | Deposit date: | 2011-06-15 | | Release date: | 2011-10-26 | | Last modified: | 2024-11-06 | | Method: | SOLID-STATE NMR | | Cite: | High-resolution membrane protein structure by joint calculations with solid-state NMR and X-ray experimental data.
J.Biomol.Nmr, 51, 2011
|
|
2LEH
 
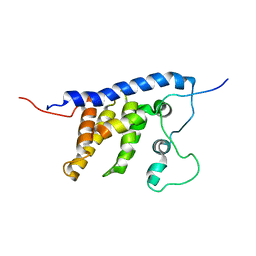 | | Solution structure of the core SMN-Gemin2 complex | | Descriptor: | Survival motor neuron protein, Survival of motor neuron protein-interacting protein 1 | | Authors: | Sarachan, K.L, Valentine, K, Gupta, K, Moorman, V, Gledhill, J, Bernens, M, Tommos, C, Wand, A.J, Van Duyne, G. | | Deposit date: | 2011-06-15 | | Release date: | 2012-06-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the core SMN-Gemin2 complex.
Biochem.J., 445, 2012
|
|
2LEJ
 
 | | human prion protein mutant HuPrP(90-231, M129, V210I) | | Descriptor: | Major prion protein | | Authors: | Biljan, I, Ilc, G, Giachin, G, Raspadori, A, Zhukov, I, Plavec, J, Legname, G. | | Deposit date: | 2011-06-16 | | Release date: | 2011-08-17 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Toward the Molecular Basis of Inherited Prion Diseases: NMR Structure of the Human Prion Protein with V210I Mutation.
J.Mol.Biol., 412, 2011
|
|
2LEK
 
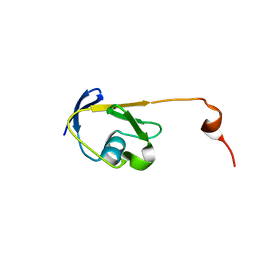 | | Solution NMR structure of a Thiamine Biosynthesis (ThiS) Protein RPA3574 from Rhodopseudomonas palustris refined with NH RDCs. Northeast Structural Genomics Consortium target RpR325 | | Descriptor: | Putative thiamin biosynthesis ThiS | | Authors: | Ramelot, T.A, Cort, J.R, Lee, H, Wang, H, Ciccosanti, C, Jiang, M, Nair, R, Rost, B, Acton, T.B, Xiao, R, Swapna, G, Everett, J.K, Prestegard, J.H, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-06-16 | | Release date: | 2011-06-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of a Thiamine Biosynthesis (ThiS) Protein RPA3574 from Rhodopseudomonas palustris. Northeast Structural Genomics Consortium target RpR325
To be Published
|
|
2LEL
 
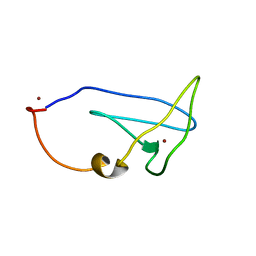 | | Structure of Cu(I)Cu(II)-CopK from Cupriavidus metallidurans CH34 | | Descriptor: | COPPER (I) ION, COPPER (II) ION, Copper resistance protein K | | Authors: | Hinds, M.G, Xiao, Z, Chong, L.X, Wedd, A.G. | | Deposit date: | 2011-06-16 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of the cooperative binding of Cu(I) and Cu(II) to the CopK protein from Cupriavidus metallidurans CH34.
Biochemistry, 50, 2011
|
|
2LEM
 
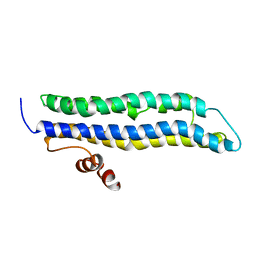 | |
2LEN
 
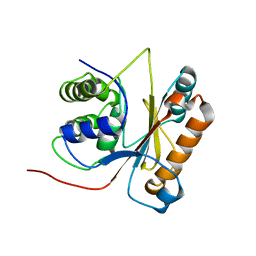 | | Solution structure of UCHL1 S18Y variant | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase isozyme L1 | | Authors: | Tse, H. | | Deposit date: | 2011-06-19 | | Release date: | 2012-06-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Backbone and side-chain (1)H, (15)N and (13)C resonance assignments of S18Y mutant of ubiquitin carboxy-terminal hydrolase L1
to be published
|
|
2LEO
 
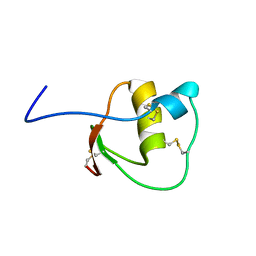 | |
2LEP
 
 | |
2LEQ
 
 | | Chemical Shift Assignment and Solution Structure of ChR145 from Cytophaga Hutchinsonii, Northeast Structural Genomics Consortium Target ChR145 | | Descriptor: | Uncharacterized protein | | Authors: | Lee, H, Lee, D, Ciccosanti, C, Mao, L.R, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Prestegard, J.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-06-21 | | Release date: | 2011-08-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of ChR145.
To be Published
|
|
2LER
 
 | | Conotoxin pc16a | | Descriptor: | Conotoxin pc16a | | Authors: | Dyubankova, N, Lescrinier, E, Van Der Haegen, A, Peigneur, S, Tytgat, J. | | Deposit date: | 2011-06-22 | | Release date: | 2012-04-11 | | Last modified: | 2024-11-27 | | Method: | SOLUTION NMR | | Cite: | Pc16a, the first characterized peptide from Conus pictus venom, shows a novel disulfide connectivity.
Peptides, 34, 2012
|
|
2LEV
 
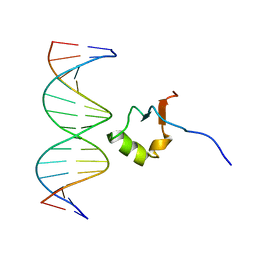 | | Structure of the DNA complex of the C-Terminal domain of Ler | | Descriptor: | DNA (5'-D(*CP*CP*TP*AP*TP*CP*AP*AP*TP*TP*AP*TP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*AP*TP*AP*AP*TP*TP*GP*AP*TP*AP*GP*G)-3'), Ler | | Authors: | Cordeiro, T.N, Schimdt, H, Madrid, C, Juarez, A, Bernado, P, Grisienger, C, Garcia, J, Pons, M. | | Deposit date: | 2011-06-23 | | Release date: | 2011-12-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Indirect DNA Readout by an H-NS Related Protein: Structure of the DNA Complex of the C-Terminal Domain of Ler.
Plos Pathog., 7, 2011
|
|
2LEW
 
 | | Structural Plasticity of Paneth cell alpha-Defensins: Characterization of Salt-Bridge Deficient Analogues of Mouse Cryptdin-4 | | Descriptor: | Alpha-defensin 4 | | Authors: | Rosengren, K, Andersson, H.S, Haugaard-Kedstrom, L.M, Bengtsson, E, Daly, N.L, Craik, D.J. | | Deposit date: | 2011-06-24 | | Release date: | 2012-05-16 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The alpha-defensin salt-bridge induces backbone stability to facilitate folding and confer proteolytic resistance.
Amino Acids, 43, 2012
|
|
2LEX
 
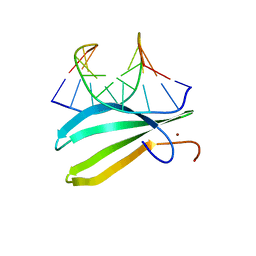 | | Complex of the C-terminal WRKY domain of AtWRKY4 and a W-box DNA | | Descriptor: | DNA (5'-D(*CP*G*CP*CP*TP*TP*TP*GP*AP*CP*CP*AP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*C*TP*GP*GP*TP*CP*AP*AP*AP*GP*GP*CP*G)-3'), Probable WRKY transcription factor 4, ... | | Authors: | Yamasaki, K, Kigawa, T, Watanabe, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-06-24 | | Release date: | 2012-01-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for sequence-spscific DNA recognition by an Arabidopsis WRKY transcription factor
J.Biol.Chem., 2012
|
|
2LEY
 
 | | Solution structure of (R7G)-Crp4 | | Descriptor: | Alpha-defensin 4 | | Authors: | Rosengren, K, Andersson, H.S, Haugaard-Kedstrom, L.M, Bengtsson, E, Daly, N.L, Figueredo, S.M, Qu, X, Craik, D.J, Ouellette, A.J. | | Deposit date: | 2011-06-26 | | Release date: | 2012-05-16 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | The alpha-defensin salt-bridge induces backbone stability to facilitate folding and confer proteolytic resistance.
Amino Acids, 43, 2012
|
|
2LEZ
 
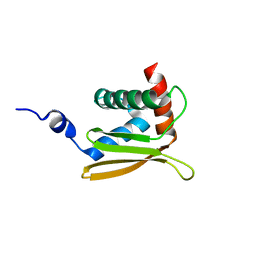 | | Solution NMR structure of N-terminal domain of Salmonella effector protein PipB2. Northeast structural genomics consortium (NESG) target stt318a | | Descriptor: | Secreted effector protein pipB2 | | Authors: | Lemak, A, Yee, A, Houliston, S, Garcia, M, Daniels, C, Savchenko, A, Arrowsmith, C, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-06-27 | | Release date: | 2011-07-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of N-terminal domain of Salmonella effector protein PipB2
To be Published
|
|
2LF0
 
 | | Solution structure of sf3636, a two-domain unknown function protein from Shigella flexneri 2a, determined by joint refinement of NMR, residual dipolar couplings and small-angle X-ray scattering, NESG target SfR339/OCSP target sf3636 | | Descriptor: | Uncharacterized protein yibL | | Authors: | Wu, B, Lemak, A, Yee, A, Lee, H, Gutmanas, A, Semesi, A, Garcia, M, Fang, X, Wang, Y, Prestegard, J.H, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2011-06-27 | | Release date: | 2011-07-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Solution structure of sf3636, a two-domain unknown function protein from Shigella flexneri 2a, determined by joint refinement of NMR, residual dipolar couplings and small-angle X-ray scattering
To be Published
|
|
2LF1
 
 | | Solution structure of L. casei dihydrofolate reductase complexed with NADPH, 30 structures | | Descriptor: | Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Polshakov, V, Feeney, J, Birdsall, B, Kovalevskaya, N. | | Deposit date: | 2011-06-28 | | Release date: | 2011-12-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structures of apo L. casei dihydrofolate reductase and its complexes with trimethoprim and NADPH: contributions to positive cooperative binding from ligand-induced refolding, conformational changes, and interligand hydrophobic interactions
Biochemistry, 50, 2011
|
|
2LF2
 
 | | Solution NMR structure of the AHSA1-like protein CHU_1110 from Cytophaga hutchinsonii, Northeast Structural Genomics Consortium Target ChR152 | | Descriptor: | Uncharacterized protein | | Authors: | Yang, Y, Ramelot, T.A, Lee, D, Ciccosanti, C, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-06-28 | | Release date: | 2011-07-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the AHSA1-like protein CHU_1110 from Cytophaga hutchinsonii, Northeast Structural Genomics Consortium Target ChR152.
To be Published
|
|
2LF3
 
 | | Solution NMR structure of HopPmaL_281_385 from Pseudomonas syringae pv. maculicola str. ES4326, Midwest Center for Structural Genomics target APC40104.5 and Northeast Structural Genomics Consortium target PsT2A | | Descriptor: | Effector protein hopAB3 | | Authors: | Wu, B, Yee, A, Houliston, S, Semesi, A, Garcia, M, Singer, A.U, Savchenko, A, Montelione, G.T, Joachimiak, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Midwest Center for Structural Genomics (MCSG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2011-06-28 | | Release date: | 2011-07-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of HopPmaL Reveals the Presence of a Second Adaptor Domain Common to the HopAB Family of Pseudomonas syringae Type III Effectors.
Biochemistry, 51, 2012
|
|
2LF4
 
 | |
2LF6
 
 | | Solution NMR structure of HopABPph1448_220_320 from Pseudomonas syringae pv. phaseolicola str. 1448A, Midwest Center for Structural Genomics target APC40132.4 and Northeast Structural Genomics Consortium target PsT3A | | Descriptor: | Effector protein hopAB1 | | Authors: | Wu, B, Yee, A, Houliston, S, Semesi, A, Garcia, M, Singer, A.U, Savchenko, A, Montelione, G.T, Joachimiak, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Midwest Center for Structural Genomics (MCSG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2011-06-28 | | Release date: | 2011-07-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of HopPmaL Reveals the Presence of a Second Adaptor Domain Common to the HopAB Family of Pseudomonas syringae Type III Effectors.
Biochemistry, 51, 2012
|
|
2LF7
 
 | |
2LF8
 
 | |
