7QUU
 
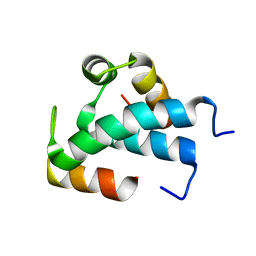 | | Red1-Iss10 complex | | Descriptor: | NURS complex subunit red1, Uncharacterized protein C7D4.14c | | Authors: | Mackereth, C.D, Kadlec, J, Laroussi, H. | | Deposit date: | 2022-01-18 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of Red1 as a conserved scaffold of the RNA-targeting MTREC/PAXT complex.
Nat Commun, 13, 2022
|
|
7QXJ
 
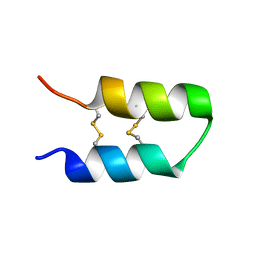 | |
7QYI
 
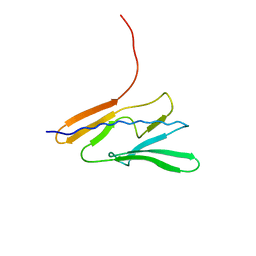 | |
7QZV
 
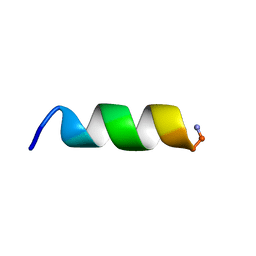 | | Hm-AMP2 | | Descriptor: | Hm-AMP2 | | Authors: | Grafskaia, E.N, Pavlova, E.R, Latsis, I.A, Malakhova, M.V, Lavrenova, V.N, Ivchenkov, D.V, Bashkirov, P.V, Kot, E.F, Mineev, K.S, Arseniev, A.S, Klinov, D.V, Lazarev, V.N. | | Deposit date: | 2022-02-01 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Non-toxic antimicrobial peptide Hm-AMP2 from leech metagenome proteins identified by the gradient-boosting approach
Materials, 224, 2022
|
|
7QZW
 
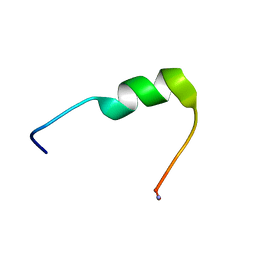 | | Hm-AMP8 | | Descriptor: | Hm-AMP8 | | Authors: | Grafskaia, E.N, Pavlova, E.R, Latsis, I.A, Malakhova, M.V, Lavrenova, V.N, Ivchenkov, D.V, Bashkirov, P.V, Kot, E.F, Mineev, K.S, Arseniev, A.S, Klinov, D.V, Lazarev, V.N. | | Deposit date: | 2022-02-01 | | Release date: | 2023-02-15 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | The study of the interaction of cationic peptides from a medical leech with blood components as a first step to the systemic use of antimicrobial peptides for therapy of infectious diseases
To Be Published
|
|
7R0R
 
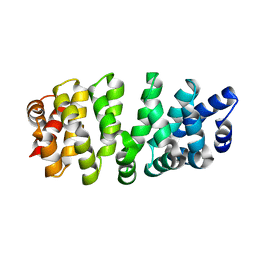 | |
7R3M
 
 | |
7R67
 
 | | Human obscurin Ig13 | | Descriptor: | Obscurin | | Authors: | Moncure, G.E, Wright, N.T. | | Deposit date: | 2021-06-22 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The N-terminus of obscurin is flexible in solution.
Proteins, 91, 2023
|
|
7R68
 
 | | Human obscurin Ig12 | | Descriptor: | Obscurin | | Authors: | Mauriello, G.E, Wright, N.T. | | Deposit date: | 2021-06-22 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The N-terminus of obscurin is flexible in solution.
Proteins, 91, 2023
|
|
7R6P
 
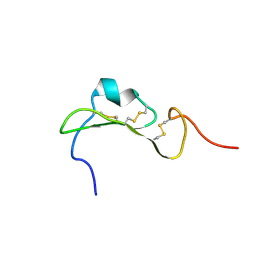 | |
7R7P
 
 | | Immature HIV-1 CACTD-SP1 lattice with Bevirimat (BVM) and Inositol hexakisphosphate (IP6) | | Descriptor: | 3alpha-[(3-carboxy-3-methylbutanoyl)oxy]-8alpha,9beta,10alpha,13alpha,17alpha,19beta-lup-20(29)-en-28-oic acid, Gag polyprotein, INOSITOL HEXAKISPHOSPHATE | | Authors: | Sarkar, S, Zadrozny, K.K, Zadorozhnyi, R, Russell, R.W, Quinn, C.M, Kleinpeter, A, Ablan, S, Meshkin, H, Perilla, J.R, Ganser-Pornillos, B.K, Pornillos, O, Freed, E.O, Gronenborn, A.M, Polenova, T. | | Deposit date: | 2021-06-25 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Structural basis of HIV-1 maturation inhibitor binding and activity.
Nat Commun, 14, 2023
|
|
7R7Q
 
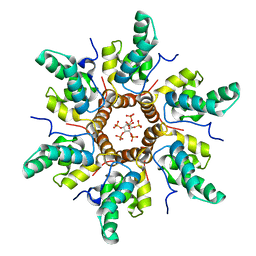 | | Immature HIV-1 CACTD-SP1 lattice with Inositol hexakisphosphate (IP6) | | Descriptor: | Gag polyprotein, INOSITOL HEXAKISPHOSPHATE | | Authors: | Sarkar, S, Zadrozny, K.K, Zadorozhnyi, R, Russell, R.W, Quinn, C.M, Kleinpeter, A, Ablan, S, Meshkin, H, Perilla, J.R, Ganser-Pornillos, B.K, Pornillos, O, Freed, E.O, Gronenborn, A.M, Polenova, T. | | Deposit date: | 2021-06-25 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Structural basis of HIV-1 maturation inhibitor binding and activity.
Nat Commun, 14, 2023
|
|
7R95
 
 | |
7RAP
 
 | |
7RC7
 
 | | Solution NMR Structure of [Ala19]Crp4 | | Descriptor: | Alpha-defensin 4 | | Authors: | Conibear, A.C, Rosengren, K.J. | | Deposit date: | 2021-07-07 | | Release date: | 2021-08-11 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | A conserved beta-bulge glycine residue facilitates folding and increases stability of the mouse alpha-defensin cryptdin-4
Peptide Science, 114, 2022
|
|
7RC8
 
 | | Solution NMR Structure of [D-Ala19]Crp4 | | Descriptor: | Alpha-defensin 4 | | Authors: | Conibear, A.C, Rosengren, K.J. | | Deposit date: | 2021-07-07 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | A conserved beta-bulge glycine residue facilitates folding and increases stability of the mouse alpha-defensin cryptdin-4
Peptide Science, 114, 2022
|
|
7RFA
 
 | |
7RIK
 
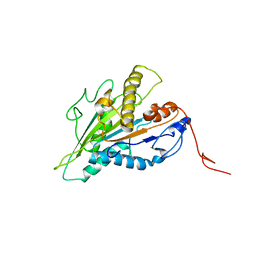 | | Magic-Angle-Spinning NMR Structure of Kinesin-1 Motor Domain Assembled with Microtubules | | Descriptor: | Kinesin-1 heavy chain | | Authors: | Zhang, C, Guo, C, Russell, R.W, Quinn, C.M, Li, M, Williams, J.C, Gronenborn, A.M, Polenova, T. | | Deposit date: | 2021-07-20 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Magic-angle-spinning NMR structure of the kinesin-1 motor domain assembled with microtubules reveals the elusive neck linker orientation
Nat Commun, 13, 2022
|
|
7RM8
 
 | |
7RN3
 
 | | hyen D solution structure | | Descriptor: | Cyclotide hyen-D | | Authors: | Du, Q, Huang, Y.H, Craik, D.J, Wang, C.K. | | Deposit date: | 2021-07-29 | | Release date: | 2022-03-02 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Mutagenesis of bracelet cyclotide hyen D reveals functionally and structurally critical residues for membrane binding and cytotoxicity.
J.Biol.Chem., 298, 2022
|
|
7RNO
 
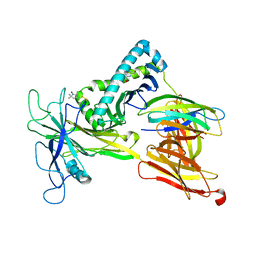 | | Model of the Ac-6-FP/hpMR1/bB2m/TAPBPR complex from integrated docking, NMR and restrained MD | | Descriptor: | Beta-2-microglobulin, Major histocompatibility complex class I-related gene protein, N-(6-formyl-4-oxo-3,4-dihydropteridin-2-yl)acetamide, ... | | Authors: | McShan, A.C, Sgourakis, N.G. | | Deposit date: | 2021-07-29 | | Release date: | 2022-05-11 | | Last modified: | 2022-08-10 | | Method: | SOLUTION NMR | | Cite: | TAPBPR employs a ligand-independent docking mechanism to chaperone MR1 molecules.
Nat.Chem.Biol., 18, 2022
|
|
7RPM
 
 | |
7RQ5
 
 | |
7RSC
 
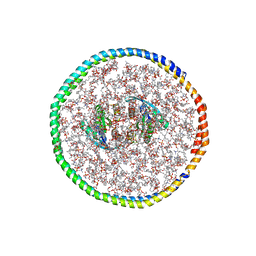 | | NMR-driven structure of the KRAS4B-G12D "alpha-alpha" dimer on a lipid bilayer nanodisc | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Lee, K, Enomoto, M, Gebregiworgis, T, Gasmi-Seabrook, G.M, Ikura, M, Marshall, C.B. | | Deposit date: | 2021-08-11 | | Release date: | 2021-09-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Oncogenic KRAS G12D mutation promotes dimerization through a second, phosphatidylserine-dependent interface: a model for KRAS oligomerization.
Chem Sci, 12, 2021
|
|
7RSE
 
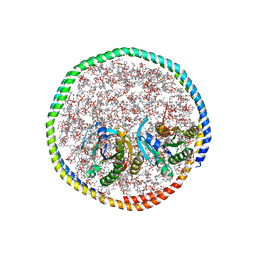 | | NMR-driven structure of the KRAS4B-G12D "alpha-beta" dimer on a lipid bilayer nanodisc | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Lee, K, Enomoto, M, Gebregiworgis, T, Gasmi-Seabrook, G.M, Ikura, M, Marshall, C.B. | | Deposit date: | 2021-08-11 | | Release date: | 2021-09-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Oncogenic KRAS G12D mutation promotes dimerization through a second, phosphatidylserine-dependent interface: a model for KRAS oligomerization.
Chem Sci, 12, 2021
|
|
