6NM2
 
 | | NMR Structure of WW291 | | Descriptor: | WW291 peptide | | Authors: | Wang, G, Zarena, D. | | Deposit date: | 2019-01-10 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The pi Configuration of the WWW Motif of a Short Trp-Rich Peptide Is Critical for Targeting Bacterial Membranes, Disrupting Preformed Biofilms, and Killing Methicillin-Resistant Staphylococcus aureus.
Biochemistry, 56, 2017
|
|
6NM3
 
 | | NMR structure of WW295 | | Descriptor: | WW295 peptide | | Authors: | Wang, G, Zarena, D. | | Deposit date: | 2019-01-10 | | Release date: | 2020-07-15 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Two distinct amphipathic peptide antibiotics with systemic efficacy.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6NNB
 
 | |
6NOA
 
 | |
6NOM
 
 | | NMR solution structure of Pisum sativum defensin 2 (Psd2) provides evidence for the presence of hydrophobic surface clusters | | Descriptor: | Defensin-2 | | Authors: | Pinheiro-Aguiar, R, Amaral, V.S.G, Bastos, I, Kurtenbach, E, Almeida, F.C.L. | | Deposit date: | 2019-01-16 | | Release date: | 2019-08-21 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance solution structure of Pisum sativum defensin 2 provides evidence for the presence of hydrophobic surface-clusters.
Proteins, 88, 2020
|
|
6NOX
 
 | | Solution structure of SFTI-KLK5 inhibitor | | Descriptor: | SFTI-KLK5 Peptide | | Authors: | White, A.M. | | Deposit date: | 2019-01-16 | | Release date: | 2019-04-03 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Amino Acid Scanning at P5' within the Bowman-Birk Inhibitory Loop Reveals Specificity Trends for Diverse Serine Proteases.
J. Med. Chem., 62, 2019
|
|
6NS8
 
 | | RDC-refined SOLUTION NMR STRUCTURE OF PROTEIN PF2048.1 | | Descriptor: | Uncharacterized protein | | Authors: | Daigham, N.S, Liu, G, Swapna, G.V.T, Cole, C, Valafar, H, Montelione, G.T. | | Deposit date: | 2019-01-24 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | REDCRAFT: A Computational Platform Using Residual Dipolar Coupling NMR Data for Determining Structures of Perdeuterated Proteins Without NOEs
To Be Published
|
|
6NU0
 
 | | Solution NMR structure of 1918 NS1 effector domain | | Descriptor: | Non-structural protein 1 | | Authors: | Shen, Q, Cho, J.H. | | Deposit date: | 2019-01-30 | | Release date: | 2020-01-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structure and conformational plasticity of the nonstructural protein 1 of the 1918 influenza A virus.
Biochem.Biophys.Res.Commun., 518, 2019
|
|
6NU4
 
 | | Solution structure of the Arabidopsis thaliana RALF8 peptide | | Descriptor: | Protein RALF-like 8 | | Authors: | Lee, W, Markley, J.L, Frederick, R.O, Miyoshi, H, Tonelli, M, Cornilescu, G, Cornilescu, C, Sussman, M.R. | | Deposit date: | 2019-01-30 | | Release date: | 2019-05-08 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Function and solution structure of the Arabidopsis thaliana RALF8 peptide.
Protein Sci., 28, 2019
|
|
6NUG
 
 | | hGRNA4-28_3s | | Descriptor: | Granulin-4 | | Authors: | Dastpeyman, M, Bansal, P, Wilson, D, Loukas, A, Smout, M, Daly, N. | | Deposit date: | 2019-02-01 | | Release date: | 2020-02-05 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | hGRN4-28_3s
to be published
|
|
6NUI
 
 | | Human Guanylate Kinase | | Descriptor: | Guanylate kinase | | Authors: | Sabo, T.M, Khan, N, Ban, D, Trigo-Mourino, P, Carneiro, M.G, Trent, J.O, Konrad, M, Lee, D. | | Deposit date: | 2019-02-01 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and functional investigation of human guanylate kinase reveals allosteric networking and a crucial role for the enzyme in cancer.
J.Biol.Chem., 294, 2019
|
|
6NVZ
 
 | |
6NW8
 
 | | SOLUTION STRUCTURE OF CN29, A TOXIN FROM CENTRUROIDES NOXIUS SCORPION VENOM | | Descriptor: | Cn29 | | Authors: | Delepierre, M, Gurrola, G.B, Possani, L.D, Guijarro, J.I. | | Deposit date: | 2019-02-06 | | Release date: | 2019-07-03 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Cn29, a novel orphan peptide found in the venom of the scorpion Centruroides noxius: Structure and function.
Toxicon, 167, 2019
|
|
6NX4
 
 | | Structure of the C-terminal Helical Repeat Domain of Eukaryotic Elongation Factor 2 Kinase (eEF-2K) | | Descriptor: | Eukaryotic elongation factor 2 kinase | | Authors: | Piserchio, A, Will, N, Giles, D.H, Hajredini, F, Dalby, K.N, Ghose, R. | | Deposit date: | 2019-02-08 | | Release date: | 2019-05-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Carboxy-Terminal Tandem Repeat Domain of Eukaryotic Elongation Factor 2 Kinase and Its Role in Substrate Recognition.
J.Mol.Biol., 431, 2019
|
|
6NZ2
 
 | |
6NZL
 
 | |
6NZN
 
 | | Dimer-of-dimer amyloid fibril structure of glucagon | | Descriptor: | Glucagon | | Authors: | Gelenter, M.D, Smith, K.J, Liao, S.Y, Mandala, V.S, Dregni, A.J, Lamm, M.S, Tian, Y, Wei, X, Pochan, D.J, Tucker, T.J, Su, Y, Hong, M. | | Deposit date: | 2019-02-14 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | The peptide hormone glucagon forms amyloid fibrils with two coexisting beta-strand conformations.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6O0C
 
 | | NMR ensemble of computationally designed protein XAA_GVDQ mutant M4L | | Descriptor: | Design construct XAA_GVDQ mutant M4L | | Authors: | Wei, K.Y, Moschidi, D, Nerli, S, Sgourakis, N, Baker, D. | | Deposit date: | 2019-02-15 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Computational design of closely related proteins that adopt two well-defined but structurally divergent folds.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6O0I
 
 | | NMR ensemble of computationally designed protein XAA | | Descriptor: | Design construct XAA | | Authors: | Wei, K.Y, Moschidi, D, Nerli, S, Sgourakis, N, Baker, D. | | Deposit date: | 2019-02-16 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Computational design of closely related proteins that adopt two well-defined but structurally divergent folds.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6O1Q
 
 | |
6O22
 
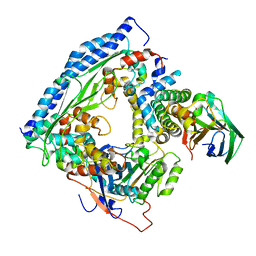 | | Structure of Asf1-H3:H4-Rtt109-Vps75 histone chaperone-lysine acetyltransferase complex with the histone substrate. | | Descriptor: | Histone H3.2, Histone H4, Histone acetyltransferase RTT109, ... | | Authors: | Danilenko, N, Carlomagno, T, Kirkpatrick, J.P. | | Deposit date: | 2019-02-22 | | Release date: | 2019-07-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Histone chaperone exploits intrinsic disorder to switch acetylation specificity.
Nat Commun, 10, 2019
|
|
6O2L
 
 | |
6O3Q
 
 | |
6O3S
 
 | | NMR solution structure of Luffin P1 | | Descriptor: | Ribosome-inactivating protein luffin P1 | | Authors: | Rosengren, K.J, Payne, C. | | Deposit date: | 2019-02-27 | | Release date: | 2019-04-24 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | An Ancient Peptide Family Buried within Vicilin Precursors.
Acs Chem.Biol., 14, 2019
|
|
6O6I
 
 | |
