2KYV
 
 | | Hybrid solution and solid-state NMR structure ensemble of phospholamban pentamer | | Descriptor: | Phospholamban | | Authors: | Verardi, R, Shi, L, Traaseth, N.J, Veglia, G. | | Deposit date: | 2010-06-08 | | Release date: | 2011-05-11 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR, SOLUTION NMR | | Cite: | Structural topology of phospholamban pentamer in lipid bilayers by a hybrid solution and solid-state NMR method.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2KYW
 
 | | Solution NMR Structure of a domain of adhesion exoprotein from Pediococcus pentosaceus, Northeast Structural Genomics Consortium Target PtR41O | | Descriptor: | Adhesion exoprotein | | Authors: | He, Y, Eletsky, A, Mills, J.L, Wang, H, Ciccosanti, C, Janjua, H, Acton, T.B, Xiao, R, Everett, J.K, Lee, H.-W, Prestegard, J.H, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-09 | | Release date: | 2010-08-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of a domain of adhesion exoprotein from Pediococcus pentosaceus, Northeast Structural Genomics Consortium Target PtR41O
To be Published
|
|
2KYX
 
 | | Solution structure of the RRM domain of CYP33 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase E | | Authors: | Park, S, Bushweller, J.H. | | Deposit date: | 2010-06-09 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The PHD3 domain of MLL acts as a CYP33-regulated switch between MLL-mediated activation and repression .
Biochemistry, 49, 2010
|
|
2KYY
 
 | | Solution NMR Structure of the N-terminal Domain of Putative ATP-dependent DNA Helicase RecG-related Protein from Nitrosomonas europaea, Northeast Structural Genomics Consortium Target NeR70A | | Descriptor: | Possible ATP-dependent DNA helicase RecG-related protein | | Authors: | Eletsky, A, Mills, J.L, Lee, H, Maglaqui, M, Ciccosanti, C, Hamilton, K, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Prestegard, J.H, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-09 | | Release date: | 2010-08-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the N-terminal Domain of Putative ATP-dependent DNA Helicase RecG-related Protein from Nitrosomonas europaea
To be Published
|
|
2KYZ
 
 | | NMR structure of heavy metal binding protein TM0320 from Thermotoga maritima | | Descriptor: | Heavy metal binding protein | | Authors: | Jaudzems, K, Wahab, A, Serrano, P, Geralt, M, Wuthrich, K, Wilson, I.A, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2010-06-09 | | Release date: | 2010-07-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure of heavy metal binding protein TM0320 from Thermotoga maritima
To be Published
|
|
2KZ0
 
 | |
2KZ1
 
 | | Inter-molecular interactions in a 44 kDa interferon-receptor complex detected by asymmetric back-protonation and 2D NOESY | | Descriptor: | Interferon alpha-2, Soluble IFN alpha/beta receptor | | Authors: | Nudelman, I, Akabayov, S.R, Schnur, E, Biron, Z, Levy, R, Xu, Y, Yang, D, Anglister, J. | | Deposit date: | 2010-06-10 | | Release date: | 2010-06-23 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Intermolecular interactions in a 44 kDa interferon-receptor complex detected by asymmetric reverse-protonation and two-dimensional NOESY
Biochemistry, 49, 2010
|
|
2KZ2
 
 | | Calmodulin, C-terminal domain, F92E mutant | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Korendovych, I, Kulp, D, Wu, Y, Cheng, H, Roder, H, DeGrado, W. | | Deposit date: | 2010-06-10 | | Release date: | 2011-04-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Design of a switchable eliminase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2KZ3
 
 | |
2KZ4
 
 | | Solution structure of protein SF1141 from Shigella flexneri 2a, Northeast structural genomics consortium (NESG) target SFT2 | | Descriptor: | Putative head-tail adaptor | | Authors: | Lemak, A, Yee, A, Garcia, M, Lee, H.-W, Prestegard, J, Montelione, G.T, Arrowsmith, C, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-11 | | Release date: | 2010-07-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of protein SF1141 from Shigella flexneri 2a
To be Published
|
|
2KZ5
 
 | | Solution NMR Structure of Transcription factor NF-E2 subunit's DNA binding domain from Homo sapiens, Northeast Structural Genomics Consortium Target HR4653B | | Descriptor: | Transcription factor NF-E2 45 kDa subunit | | Authors: | Liu, G, Janjua, H, Xiao, R, Ciccosanti, C, Shastry, R, Acton, T.B, Tong, S, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-11 | | Release date: | 2010-07-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Northeast Structural Genomics Consortium Target HR4653B
To be Published
|
|
2KZ6
 
 | | Solution structure of protein CV0426 from Chromobacterium violaceum, Northeast structural genomics consortium (NESG) target CVT2 | | Descriptor: | Uncharacterized protein | | Authors: | Lemak, A, Yee, A, Lee, H, Semesi, A, Prestegard, J, Montelione, G.T, Arrowsmith, C, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-11 | | Release date: | 2010-06-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of protein CV0426 from Chromobacterium violaceum
To be Published
|
|
2KZ7
 
 | |
2KZ8
 
 | | Solution NMR structure of MqsA, a protein from E. coli, containing a Zinc finger, N-terminal and a Helix Turn-Helix C-terminal domain | | Descriptor: | Uncharacterized HTH-type transcriptional regulator ygiT | | Authors: | Papadopoulos, E, Vlamis-Gardikas, A, Graslund, A, Billeter, M, Holmgren, A, Collet, J. | | Deposit date: | 2010-06-14 | | Release date: | 2011-06-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and biophysical properties of MqsA, a Zn-containing antitoxin from Escherichia coli
Biochim.Biophys.Acta, 2012
|
|
2KZ9
 
 | |
2KZA
 
 | | Solution structure of ASIP(80-132, P103A, P105A, Q115Y, S124Y) | | Descriptor: | Agouti-signaling protein | | Authors: | Patel, M.P, Cribb Fabersunne, C.S, Yang, Y, Kaelin, C.B, Barsh, G.S, Millhauser, G.L. | | Deposit date: | 2010-06-14 | | Release date: | 2010-07-21 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Loop-swapped chimeras of the agouti-related protein and the agouti signaling protein identify contacts required for melanocortin 1 receptor selectivity and antagonism.
J.Mol.Biol., 404, 2010
|
|
2KZB
 
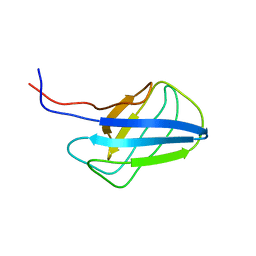 | | Solution structure of alpha-mannosidase binding domain of Atg19 | | Descriptor: | Autophagy-related protein 19 | | Authors: | Watanabe, Y, Noda, N, Kumeta, H, Suzuki, K, Ohsumi, Y, Inagaki, F. | | Deposit date: | 2010-06-15 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Selective transport of alpha-mannosidase by autophagic pathways: structural basis for cargo recognition by Atg19 and Atg34.
J.Biol.Chem., 285, 2010
|
|
2KZC
 
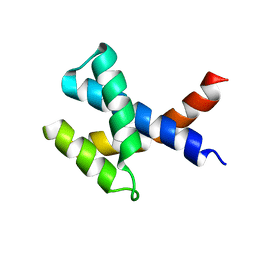 | | Solution NMR structure of the protein YP_510488.1 | | Descriptor: | Uncharacterized protein | | Authors: | Mohanty, B, Serrano, P, Geralt, M, Horst, R, Wilson, I.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2010-06-15 | | Release date: | 2010-07-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the protein YP_510488.1
To be Published
|
|
2KZD
 
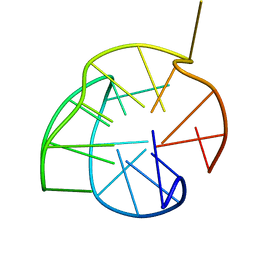 | | Structure of a (3+1) G-quadruplex formed by hTERT promoter sequence | | Descriptor: | DNA (5'-D(*AP*GP*GP*GP*IP*AP*GP*GP*GP*GP*CP*TP*GP*GP*GP*AP*GP*GP*GP*C)-3') | | Authors: | Lim, K.W, Lacroix, L, Yue, D.J.E, Lim, J.K.C, Lim, J.M.W, Phan, A.T. | | Deposit date: | 2010-06-16 | | Release date: | 2010-10-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Coexistence of two distinct G-quadruplex conformations in the hTERT promoter
J.Am.Chem.Soc., 132, 2010
|
|
2KZE
 
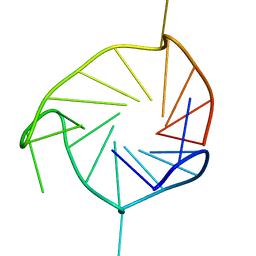 | | Structure of an all-parallel-stranded G-quadruplex formed by hTERT promoter sequence | | Descriptor: | DNA (5'-D(*AP*IP*GP*GP*GP*AP*GP*GP*GP*IP*CP*TP*GP*GP*GP*AP*GP*GP*GP*C)-3') | | Authors: | Lim, K.W, Lacroix, L, Yue, D.J.E, Lim, J.K.C, Lim, J.M.W, Phan, A.T. | | Deposit date: | 2010-06-16 | | Release date: | 2010-10-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Coexistence of two distinct G-quadruplex conformations in the hTERT promoter
J.Am.Chem.Soc., 132, 2010
|
|
2KZF
 
 | | Solution NMR structure of the thermotoga maritima protein TM0855 a putative ribosome binding factor A | | Descriptor: | Ribosome-binding factor A | | Authors: | Serrano, P, Jaudzems, K, Horst, R, Wilson, I.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2010-06-16 | | Release date: | 2010-08-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the thermotoga maritima protein TM0855
To be Published
|
|
2KZG
 
 | | A Transient and Low Populated Protein Folding Intermediate at Atomic Resolution | | Descriptor: | Pre-mRNA-processing factor 40 homolog A | | Authors: | Korzhnev, D.M, Religa, T.L, Banachewicz, W, Fersht, A.R, Kay, L.E. | | Deposit date: | 2010-06-17 | | Release date: | 2010-09-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A transient and low-populated protein-folding intermediate at atomic resolution.
Science, 329, 2010
|
|
2KZH
 
 | |
2KZI
 
 | | Solution structure of the ZHER2 Affibody | | Descriptor: | Engineered protein, ZHER2 Affibody | | Authors: | Hard, T. | | Deposit date: | 2010-06-18 | | Release date: | 2010-08-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for high-affinity HER2 receptor binding by an engineered protein.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2KZJ
 
 | | Solution structure of the ZHER2 Affibody (alternative) | | Descriptor: | Engineered protein, ZHER2 Affibody | | Authors: | Hard, T. | | Deposit date: | 2010-06-18 | | Release date: | 2010-08-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for high-affinity HER2 receptor binding by an engineered protein.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
