7WE3
 
 | |
7WEI
 
 | |
7WEM
 
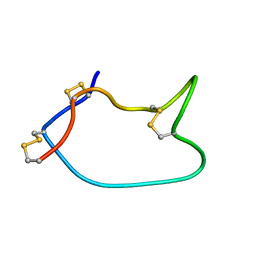
 | | Solid-state NMR Structure of TFo c-Subunit Ring | | Descriptor: | ATP synthase subunit c | | Authors: | Akutsu, H, Todokoro, Y, Kang, S.-J, Suzuki, T, Yoshida, M, Ikegami, T, Fujiwara, T. | | Deposit date: | 2021-12-23 | | Release date: | 2022-08-10 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Chemical Conformation of the Essential Glutamate Site of the c -Ring within Thermophilic Bacillus F o F 1 -ATP Synthase Determined by Solid-State NMR Based on its Isolated c -Ring Structure.
J.Am.Chem.Soc., 144, 2022
|
|
7WGW
 
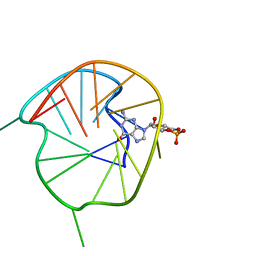 | | NMR Solution Structure of a cGMP Fill-in Vacancy G-quadruplex Formed in the Oxidized BLM Gene Promoter | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, DNA (20-MER) | | Authors: | Wang, K.B, Liu, Y, Li, Y, Li, J, Dickerhoff, J, Yang, M.H, Yang, D, Kong, L.Y. | | Deposit date: | 2021-12-29 | | Release date: | 2022-05-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Oxidative Damage Induces a Vacancy G-Quadruplex That Binds Guanine Metabolites: Solution Structure of a cGMP Fill-in Vacancy G-Quadruplex in the Oxidized BLM Gene Promoter.
J.Am.Chem.Soc., 144, 2022
|
|
7WH3
 
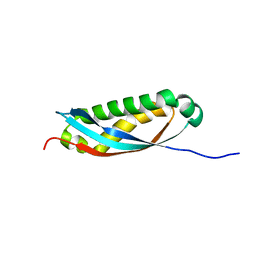 | | Solution structure of human stomatin SPFH domain in a phosphate buffer | | Descriptor: | Stomatin | | Authors: | Kataoka, K, Tenno, T, Goda, N, Hibino, E, Hiroaki, H. | | Deposit date: | 2021-12-29 | | Release date: | 2022-06-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A cryptic phosphate-binding pocket on the SPFH domain of human stomatin that regulates a novel fibril-like self-assembly
Curr Res Struct Biol, 4, 2022
|
|
7WIO
 
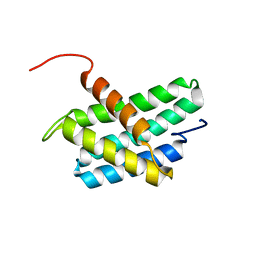 | | NMR structure of N-terminal domain of Triconephila clavipes of major ampullate spidroin 1 | | Descriptor: | Major ampullate spidroin 1A | | Authors: | Oktaviani, N.A, Malay, A.D, Matsugami, A, Hayashi, F, Numata, K. | | Deposit date: | 2022-01-04 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Unusual p K a Values Mediate the Self-Assembly of Spider Dragline Silk Proteins.
Biomacromolecules, 24, 2023
|
|
7WJ0
 
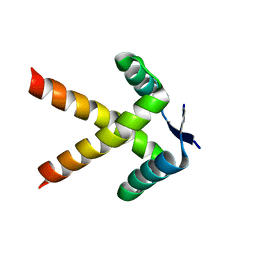 | |
7WKC
 
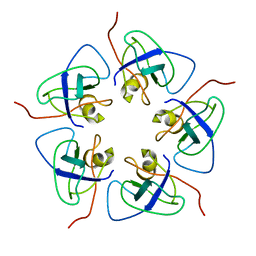 | |
7WKF
 
 | | Antimicrobial peptide-LaIT2 | | Descriptor: | Beta-KTx-like peptide LaIT2 | | Authors: | Tamura, M, Morita, H, Ohki, S. | | Deposit date: | 2022-01-09 | | Release date: | 2023-04-05 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Structural and functional studies of LaIT2, an antimicrobial and insecticidal peptide from Liocheles australasiae.
Toxicon, 214, 2022
|
|
7WNR
 
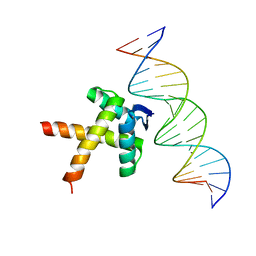 | | Data-driven HADDOCK model of mycobacterial nMazE6-operator DNA complex | | Descriptor: | Antitoxin MazE6, DNA (5'-D(P*CP*CP*GP*GP*TP*TP*AP*TP*AP*CP*TP*AP*TP*CP*TP*GP*TP*A)-3'), DNA (5'-D(P*TP*AP*CP*AP*GP*AP*TP*AP*GP*TP*AP*TP*AP*AP*CP*CP*GP*G)-3') | | Authors: | Kumari, K, Sarma, S.P. | | Deposit date: | 2022-01-19 | | Release date: | 2022-10-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and mutational analysis of MazE6-operator DNA complex provide insights into autoregulation of toxin-antitoxin systems.
Commun Biol, 5, 2022
|
|
7X2Z
 
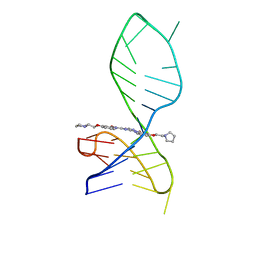 | | NMR solution structure of the 1:1 complex of a pyridostatin derivative (PyPDS) bound to a G-quadruplex MYT1L | | Descriptor: | 4-(2-azanylethoxy)-N2,N6-bis[4-(2-pyrrolidin-1-ylethoxy)quinolin-2-yl]pyridine-2,6-dicarboxamide, G-quadruplex DNA MYT1L | | Authors: | Liu, L.-Y, Mao, Z.-W, Liu, W. | | Deposit date: | 2022-02-26 | | Release date: | 2022-06-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Pyridostatin and Its Derivatives Specifically Binding to G-Quadruplexes.
J.Am.Chem.Soc., 144, 2022
|
|
7X31
 
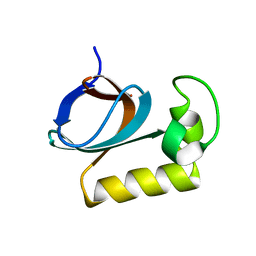 | | solution structure of an anti-CRISPR protein | | Descriptor: | Anti-CRISPR protein (AcrIIC1) | | Authors: | Zhao, Y, Yang, F. | | Deposit date: | 2022-02-27 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A redox switch regulates the assembly and anti-CRISPR activity of AcrIIC1.
Nat Commun, 13, 2022
|
|
7X3A
 
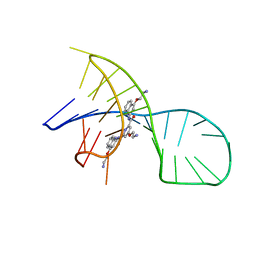 | | NMR solution structure of the 1:1 complex of a pyridostatin (PDS) bound to a G-quadruplex MYT1L | | Descriptor: | 4-(2-azanylethoxy)-N2,N6-bis[4-(2-azanylethoxy)quinolin-2-yl]pyridine-2,6-dicarboxamide, G-quadruplex DNA MYT1L | | Authors: | Liu, L.-Y, Mao, Z.-W, Liu, W. | | Deposit date: | 2022-02-28 | | Release date: | 2022-06-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Pyridostatin and Its Derivatives Specifically Binding to G-Quadruplexes.
J.Am.Chem.Soc., 144, 2022
|
|
7X5C
 
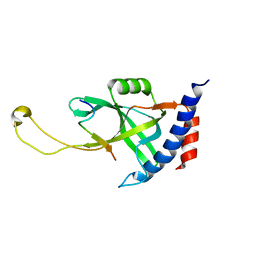 | | Solution structure of Tetrahymena p75OB1-p50PBM | | Descriptor: | Telomerase associated protein p50PBM, Telomerase-associated protein p75OB1 | | Authors: | Wu, B, Tang, T, Xue, H.J, Wu, J, Lei, M. | | Deposit date: | 2022-03-04 | | Release date: | 2022-10-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Association of the CST complex and p50 in Tetrahymena is crucial for telomere maintenance
Structure, 2022
|
|
7X7S
 
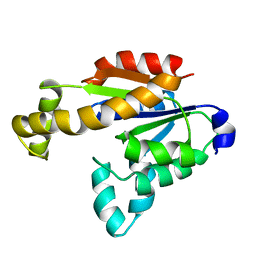 | | Solution structure of human adenylate kinase 1 (hAK1) | | Descriptor: | Adenylate kinase isoenzyme 1 | | Authors: | Zhang, H. | | Deposit date: | 2022-03-10 | | Release date: | 2022-05-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | ADP-Induced Conformational Transition of Human Adenylate Kinase 1 Is Triggered by Suppressing Internal Motion of alpha 3 alpha 4 and alpha 7 alpha 8 Fragments on the ps-ns Timescale.
Biomolecules, 12, 2022
|
|
7X89
 
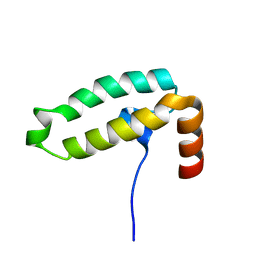 | | Tid1 | | Descriptor: | DnaJ homolog subfamily A member 3, mitochondrial | | Authors: | Jang, J, Lee, S.H, Kang, D.H, Sim, D.W, Jo, K.S, Ryu, H, Kim, E.H, Ryu, K.S, Lee, J.H, Kim, J.H, Won, H.S. | | Deposit date: | 2022-03-11 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural studies on the J-domain and GF-motif of the mitochondrial Hsp40, Tid1
To Be Published
|
|
7X8M
 
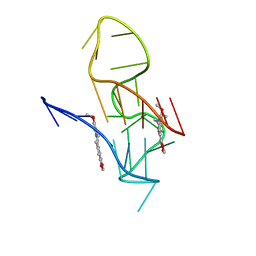 | | NMR Solution Structure of the 2:1 Berberine-KRAS-G4 Complex | | Descriptor: | BERBERINE, DNA (24-MER) | | Authors: | Wang, K.B, Liu, Y, Li, J, Xiao, C, Gu, W, Li, Y, Xia, Y.Z, Yan, T, Yang, M.H, Kong, L.Y. | | Deposit date: | 2022-03-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the bulge-containing KRAS oncogene promoter G-quadruplex bound to berberine and coptisine.
Nat Commun, 13, 2022
|
|
7X8N
 
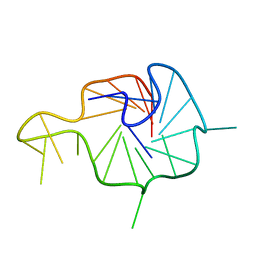 | | NMR Solution Structure of the Wild-type Bulge-containing KRAS-G4 | | Descriptor: | DNA (24-mer) | | Authors: | Wang, K.B, Liu, Y, Li, J, Xiao, C, Gu, W, Li, Y, Xia, Y.Z, Yan, T, Yang, M.H, Kong, L.Y. | | Deposit date: | 2022-03-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the bulge-containing KRAS oncogene promoter G-quadruplex bound to berberine and coptisine.
Nat Commun, 13, 2022
|
|
7X8O
 
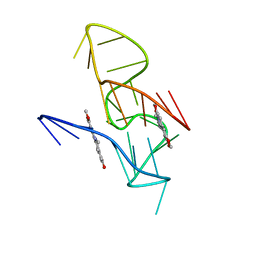 | | NMR Solution Structure of the 2:1 Coptisine-KRAS-G4 Complex | | Descriptor: | 6,7-dihydro[1,3]dioxolo[4,5-g][1,3]dioxolo[7,8]isoquino[3,2-a]isoquinolin-5-ium, DNA (24-MER) | | Authors: | Wang, K.B, Liu, Y, Li, J, Xiao, C, Gu, W, Li, Y, Xia, Y.Z, Yan, T, Yang, M.H, Kong, L.Y. | | Deposit date: | 2022-03-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the bulge-containing KRAS oncogene promoter G-quadruplex bound to berberine and coptisine.
Nat Commun, 13, 2022
|
|
7X9U
 
 | | Type-II KH motif of human mitochondrial RbfA | | Descriptor: | Putative ribosome-binding factor A, mitochondrial | | Authors: | Kuwasako, K, Suzuki, S, Furue, M, Takizawa, M, Takahashi, M, Tsuda, K, Nagata, T, Watanabe, S, Tanaka, A, Kobayashi, N, Kigawa, T, Guntert, P, Shirouzu, M, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2022-03-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1 H, 13 C, and 15 N resonance assignments and solution structures of the KH domain of human ribosome binding factor A, mtRbfA, involved in mitochondrial ribosome biogenesis.
Biomol.Nmr Assign., 16, 2022
|
|
7XEZ
 
 | |
7XFG
 
 | |
7XFV
 
 | |
7XGA
 
 | |
7XH3
 
 | |
