5NPN
 
 | |
5NQ4
 
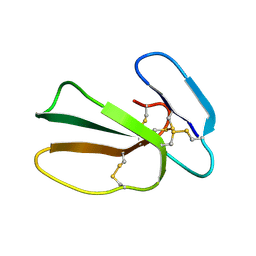 | | Cytotoxin-1 in DPC-micelle | | Descriptor: | Cytotoxin 1 | | Authors: | Dubovskii, P.V, Dubinnyi, M.A, Volynsky, P.E, Pustovalova, Y.E, Konshina, A.G, Utkin, Y.N, Efremov, R.G, Arseniev, A.S. | | Deposit date: | 2017-04-19 | | Release date: | 2017-12-13 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Impact of membrane partitioning on the spatial structure of an S-type cobra cytotoxin.
J. Biomol. Struct. Dyn., 36, 2018
|
|
5NR5
 
 | |
5NR6
 
 | |
5NVB
 
 | |
5NVP
 
 | | NMR assignment and structure of a peptide derived from the fusion peptide of HIV-1 gp41 in the presence of dodecylphosphocholine micelles | | Descriptor: | Envelope glycoprotein,Gp41 | | Authors: | Jimenez, M.A, Serrano, S, Nieva, J.L, Huarte, N. | | Deposit date: | 2017-05-04 | | Release date: | 2017-12-06 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Structure-Related Roles for the Conservation of the HIV-1 Fusion Peptide Sequence Revealed by Nuclear Magnetic Resonance.
Biochemistry, 56, 2017
|
|
5NWM
 
 | |
5NWU
 
 | | NMR assignment and structure of a peptide derived from the fusion peptide of HIV-1 gp41 in the presence of hexafluoroisopropanol | | Descriptor: | wtFP-tag,Gp41 | | Authors: | Jimenez, M.A, Serrano, S, Nieva, J.L, Huarte, N. | | Deposit date: | 2017-05-08 | | Release date: | 2017-12-06 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Structure-Related Roles for the Conservation of the HIV-1 Fusion Peptide Sequence Revealed by Nuclear Magnetic Resonance.
Biochemistry, 56, 2017
|
|
5NWV
 
 | | NMR assignment and structure of a peptide derived from the fusion peptide of HIV-1 gp41 in the presence of dodecylphosphocholine micelles | | Descriptor: | scrFP-tag,Gp41 | | Authors: | Jimenez, M.A, Serrano, S, Nieva, J.L, Huarte, N. | | Deposit date: | 2017-05-08 | | Release date: | 2017-12-06 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Structure-Related Roles for the Conservation of the HIV-1 Fusion Peptide Sequence Revealed by Nuclear Magnetic Resonance.
Biochemistry, 56, 2017
|
|
5NWW
 
 | | NMR assignment and structure of a peptide derived from the fusion peptide of HIV-1 gp41 in the presence of dodecylphosphocholine micelles | | Descriptor: | scrFP-tag,Gp41 | | Authors: | Jimenez, M.A, Serrano, S, Nieva, J.L, Huarte, N. | | Deposit date: | 2017-05-08 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structure-Related Roles for the Conservation of the HIV-1 Fusion Peptide Sequence Revealed by Nuclear Magnetic Resonance.
Biochemistry, 56, 2017
|
|
5NYS
 
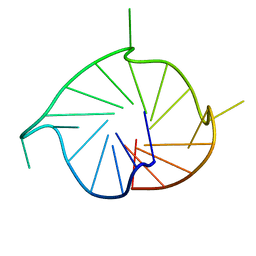 | | M2 G-quadruplex dilute solution | | Descriptor: | DNA (5'-D(*TP*AP*GP*GP*GP*AP*CP*GP*GP*GP*CP*GP*GP*GP*CP*AP*GP*GP*GP*T)-3') | | Authors: | Trajkovski, M, Plavec, J, Endoh, T, Tateishi-Karimata, H, Sugimoto, N. | | Deposit date: | 2017-05-11 | | Release date: | 2018-04-11 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Pursuing origins of (poly)ethylene glycol-induced G-quadruplex structural modulations.
Nucleic Acids Res., 46, 2018
|
|
5NYT
 
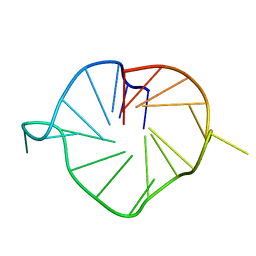 | | M2 G-quadruplex 20 wt% ethylene glycol | | Descriptor: | DNA (5'-D(*TP*AP*GP*GP*GP*AP*CP*GP*GP*GP*CP*GP*GP*GP*CP*AP*GP*GP*GP*T)-3') | | Authors: | Trajkovski, M, Plavec, J, Endoh, T, Tateishi-Karimata, H, Sugimoto, N. | | Deposit date: | 2017-05-11 | | Release date: | 2018-04-11 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Pursuing origins of (poly)ethylene glycol-induced G-quadruplex structural modulations.
Nucleic Acids Res., 46, 2018
|
|
5NYU
 
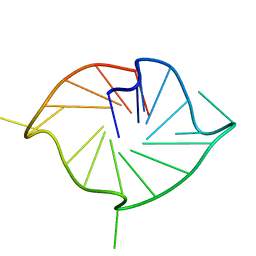 | | M2 G-quadruplex 10 wt% PEG8000 | | Descriptor: | DNA (5'-D(*TP*AP*GP*GP*GP*AP*CP*GP*GP*GP*CP*GP*GP*GP*CP*AP*GP*GP*GP*T)-3') | | Authors: | Trajkovski, M, Plavec, J, Endoh, T, Tateishi-Karimata, H, Sugimoto, N. | | Deposit date: | 2017-05-11 | | Release date: | 2018-04-11 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Pursuing origins of (poly)ethylene glycol-induced G-quadruplex structural modulations.
Nucleic Acids Res., 46, 2018
|
|
5NZ9
 
 | | NMR structure of an EphA2-Sam fragment | | Descriptor: | Ephrin type-A receptor 2 | | Authors: | Mercurio, F.A, Leone, M. | | Deposit date: | 2017-05-12 | | Release date: | 2017-12-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Sam-Sam interaction between Ship2 and the EphA2 receptor: design and analysis of peptide inhibitors.
Sci Rep, 7, 2017
|
|
5O1Q
 
 | | LysF1 sh3b domain structure | | Descriptor: | sh3b domain | | Authors: | Benesik, M, Novacek, J, Janda, L, Dopitova, R, Pernisova, M, Melkova, K, Tisakova, L, Doskar, J, Zidek, L, Hejatko, J, Pantucek, R. | | Deposit date: | 2017-05-19 | | Release date: | 2017-09-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Role of SH3b binding domain in a natural deletion mutant of Kayvirus endolysin LysF1 with a broad range of lytic activity.
Virus Genes, 54, 2018
|
|
5O1T
 
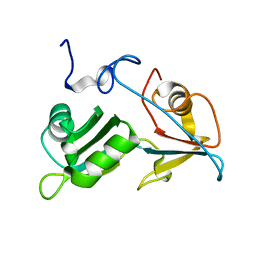 | |
5O2M
 
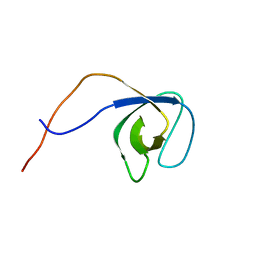 | | p130Cas SH3 domain | | Descriptor: | Breast cancer anti-estrogen resistance 1 | | Authors: | Hexnerova, R, Veverka, V. | | Deposit date: | 2017-05-22 | | Release date: | 2017-09-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of CAS SH3 domain selectivity and regulation reveals new CAS interaction partners.
Sci Rep, 7, 2017
|
|
5O2P
 
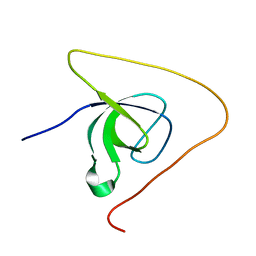 | | p130Cas SH3 domain PTP-PEST peptide chimera | | Descriptor: | Breast cancer anti-estrogen resistance 1,Tyrosine-protein phosphatase non-receptor type 12 | | Authors: | Hexnerova, R, Veverka, V. | | Deposit date: | 2017-05-22 | | Release date: | 2017-09-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of CAS SH3 domain selectivity and regulation reveals new CAS interaction partners.
Sci Rep, 7, 2017
|
|
5O2Q
 
 | | p130Cas SH3 domain Vinculin peptide chimera | | Descriptor: | Breast cancer anti-estrogen resistance 1,Vinculin | | Authors: | Hexnerova, R, Veverka, V. | | Deposit date: | 2017-05-22 | | Release date: | 2017-09-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of CAS SH3 domain selectivity and regulation reveals new CAS interaction partners.
Sci Rep, 7, 2017
|
|
5O2V
 
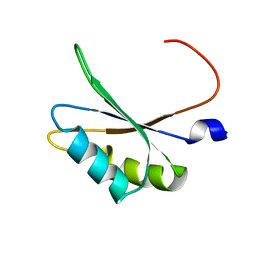 | | NMR structure of TIA-1 RRM1 domain | | Descriptor: | Nucleolysin TIA-1 isoform p40 | | Authors: | Jagtap, P.K.A. | | Deposit date: | 2017-05-22 | | Release date: | 2017-06-28 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Segmental, Domain-Selective Perdeuteration and Small-Angle Neutron Scattering for Structural Analysis of Multi-Domain Proteins.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5O2Y
 
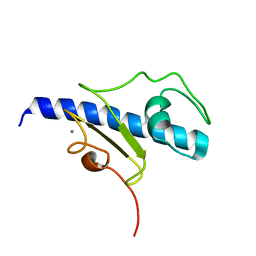 | | NMR structure of the calcium bound form of PulG, major pseudopilin from Klebsiella oxytoca T2SS | | Descriptor: | CALCIUM ION, General secretion pathway protein G | | Authors: | Lopez-Castilla, A, Bardiaux, B, Vitorge, B, Thomassin, J.-L, Zheng, W, Yu, X, Egelman, E.H, Nilges, M, Francetic, O, Izadi-Pruneyre, N. | | Deposit date: | 2017-05-23 | | Release date: | 2017-10-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the calcium-dependent type 2 secretion pseudopilus.
Nat Microbiol, 2, 2017
|
|
5O4D
 
 | | G-quadruplex of Human papillomavirus type 52 | | Descriptor: | DNA (5'-D(*GP*GP*GP*TP*AP*GP*GP*GP*CP*AP*GP*GP*GP*GP*AP*CP*AP*CP*AP*GP*GP*GP*T)-3') | | Authors: | Marusic, M, Plavec, J. | | Deposit date: | 2017-05-29 | | Release date: | 2018-06-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Towards Understanding of Polymorphism of the G-rich Region of Human Papillomavirus Type 52.
Molecules, 24, 2019
|
|
5O57
 
 | | Solution Structure of the N-terminal Region of Dkk4 | | Descriptor: | Dickkopf-related protein 4 | | Authors: | Waters, L.C, Patel, S, Barkell, A.M, Muskett, F.W, Robinson, M.K, Holdsworth, G, Carr, M.D. | | Deposit date: | 2017-06-01 | | Release date: | 2018-06-13 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Structural and functional analysis of Dickkopf 4 (Dkk4): New insights into Dkk evolution and regulation of Wnt signaling by Dkk and Kremen proteins.
J. Biol. Chem., 293, 2018
|
|
5O6F
 
 | | NMR structure of cold shock protein A from Corynebacterium pseudotuberculosis | | Descriptor: | Cold-shock protein | | Authors: | Caruso, I.P, Panwalkar, V, Coronado, M.A, Dingley, A.J, Cornelio, M.L, Willbold, D, Arni, R.K, Eberle, R.J. | | Deposit date: | 2017-06-06 | | Release date: | 2017-07-19 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure and interaction of Corynebacterium pseudotuberculosis cold shock protein A with Y-box single-stranded DNA fragment.
FEBS J., 285, 2018
|
|
5O7J
 
 | | Structural insights into the periplasmic sensor domain of the GacS histidine kinase controlling biofilm formation in Pseudomonas aeruginosa | | Descriptor: | Histidine kinase | | Authors: | Ali-Ahmad, A, Bornet, O, Fadel, F, Bourne, Y, Vincent, F, Guerlesquin, F, Sebban-Kreuzer, C. | | Deposit date: | 2017-06-09 | | Release date: | 2018-02-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural and functional insights into the periplasmic detector domain of the GacS histidine kinase controlling biofilm formation in Pseudomonas aeruginosa.
Sci Rep, 7, 2017
|
|
