2MTK
 
 | |
2MTL
 
 | | Solution NMR Structure of De novo designed FR55, Northeast Structural Genomics Consortium (NESG) Target OR109 | | Descriptor: | De novo designed protein FR55 OR109 | | Authors: | Liu, G, Koga, N, Koga, R, Xiao, R, Hamilton, K, Ciccosanti, C, Sahdev, S, Kohan, E, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-08-19 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of De novo designed FR55, Northeast Structural Genomics Consortium (NESG) Target OR109
To be Published
|
|
2MTM
 
 | | NMR structure of RCB-1 peptide | | Descriptor: | Putative uncharacterized protein | | Authors: | Boldbaatar, D, Elseedi, H.R. | | Deposit date: | 2014-08-21 | | Release date: | 2015-09-02 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Synthesis, Structural Characterization, and Bioactivity of the Stable Peptide RCB-1 from Ricinus communis.
J Nat Prod, 78, 2015
|
|
2MTN
 
 | | Solution structure of MLL-IBD complex | | Descriptor: | Histone-lysine N-methyltransferase 2A, PC4 and SFRS1-interacting protein fusion | | Authors: | Cierpicki, T, Pollock, J, Murai, M. | | Deposit date: | 2014-08-23 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The same site on the integrase-binding domain of lens epithelium-derived growth factor is a therapeutic target for MLL leukemia and HIV.
Blood, 124, 2014
|
|
2MTO
 
 | | Non-reducible analogues of alpha-conotoxin RgIA: [2,8]-cis dicarba RgIA | | Descriptor: | Alpha-conotoxin RgIA | | Authors: | Chhabra, S, Robinson, S, Norton, R. | | Deposit date: | 2014-08-26 | | Release date: | 2014-11-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Dicarba Analogues of alpha-Conotoxin RgIA. Structure, Stability, and Activity at Potential Pain Targets.
J.Med.Chem., 57, 2014
|
|
2MTP
 
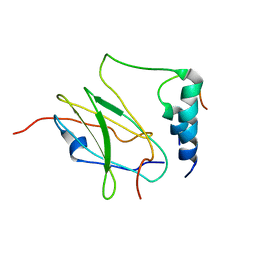 | |
2MTQ
 
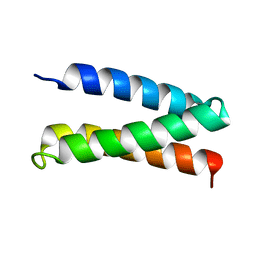 | | Solution Structure of a De Novo Designed Peptide that Sequesters Toxic Heavy Metals | | Descriptor: | Designed Peptide | | Authors: | Plegaria, J.S, Zuiderweg, E.R, Stemmler, T.L, Pecoraro, V.L. | | Deposit date: | 2014-08-28 | | Release date: | 2015-04-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Apoprotein Structure and Metal Binding Characterization of a de Novo Designed Peptide, alpha 3DIV, that Sequesters Toxic Heavy Metals.
Biochemistry, 54, 2015
|
|
2MTS
 
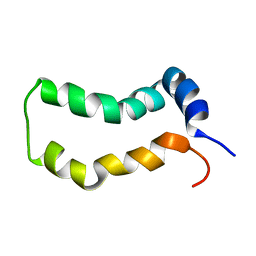 | | Three-Dimensional Structure and Interaction Studies of Hepatitis C Virus p7 in 1,2-Dihexanoyl-sn-glycero-3-phosphocholine by Solution Nuclear Magnetic Resonance | | Descriptor: | HEPATITIS C VIRUS P7 PROTEIN | | Authors: | Cook, G.A, Dawson, L.A, Tian, Y, Opella, S.J. | | Deposit date: | 2014-08-29 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure and interaction studies of hepatitis C virus p7 in 1,2-dihexanoyl-sn-glycero-3-phosphocholine by solution nuclear magnetic resonance.
Biochemistry, 52, 2013
|
|
2MTT
 
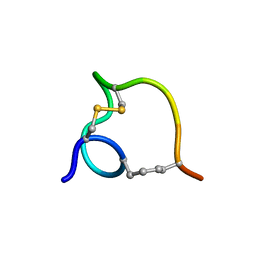 | | Non-reducible analogues of alpha-conotoxin RgIA: [3,12]-cis dicarba RgIA | | Descriptor: | Dicarba Analogues of alpha-Conotoxin RgIA | | Authors: | Chhabra, S, Robinson, S.D, Norton, R.S. | | Deposit date: | 2014-08-31 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Dicarba Analogues of alpha-Conotoxin RgIA. Structure, Stability, and Activity at Potential Pain Targets.
J.Med.Chem., 57, 2014
|
|
2MTU
 
 | | Non-reducible analogues of alpha-conotoxin RgIA: [3,12]-trans dicarba RgIA | | Descriptor: | Dicarba Analogues of alpha-Conotoxin RgIA | | Authors: | Chhabra, S, Robinson, S.D, Norton, R.S. | | Deposit date: | 2014-09-01 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Dicarba Analogues of alpha-Conotoxin RgIA. Structure, Stability, and Activity at Potential Pain Targets.
J.Med.Chem., 57, 2014
|
|
2MTV
 
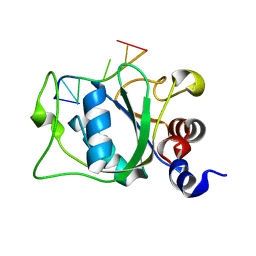 | | Solution Structure of the YTH Domain of YT521-B in complex with N6-Methyladenosine containing RNA | | Descriptor: | RNA_(5'-R(*UP*GP*(6MZ)P*CP*AP*C)-3'), YTH domain-containing protein 1 | | Authors: | Theler, D, Dominguez, C, Blatter, M, Boudet, J, Allain, F.H.-T. | | Deposit date: | 2014-09-01 | | Release date: | 2014-11-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the YTH domain in complex with N6-methyladenosine RNA: a reader of methylated RNA.
Nucleic Acids Res., 42, 2014
|
|
2MTW
 
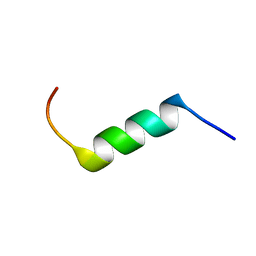 | | Evidence supporting the hypothesis that specifically modifying a malaria peptide to fit into HLA-DR 1*03 molecules induces antibody production and protection | | Descriptor: | Erythrocyte-binding antigen 175 | | Authors: | Cifuentes, G, Salazar, L, Vargas, L, Parra, C, Vanegas, M, Cortes, J, Sandoval, M, Patarroyo, M.E. | | Deposit date: | 2014-09-01 | | Release date: | 2015-02-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Evidence supporting the hypothesis that specifically modifying a malaria peptide to fit HLA-DR 1*03 molecules induces antibody production and protection
To be Published
|
|
2MTX
 
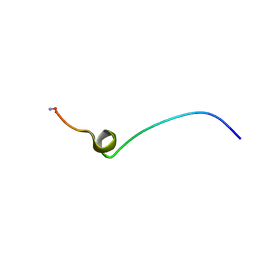 | |
2MTY
 
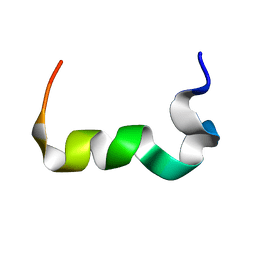 | |
2MTZ
 
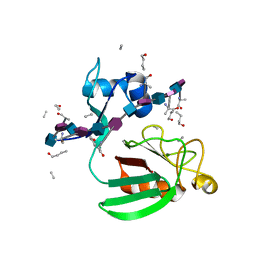 | | Haddock model of Bacillus subtilis L,D-transpeptidase in complex with a peptidoglycan hexamuropeptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid, Putative L,D-transpeptidase YkuD, intact bacterial peptidoglycan | | Authors: | Schanda, P, Triboulet, S, Laguri, C, Bougault, C, Ayala, I, Callon, M, Arthur, M, Simorre, J. | | Deposit date: | 2014-09-02 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic model of a cell-wall cross-linking enzyme in complex with an intact bacterial peptidoglycan.
J.Am.Chem.Soc., 136, 2014
|
|
2MU0
 
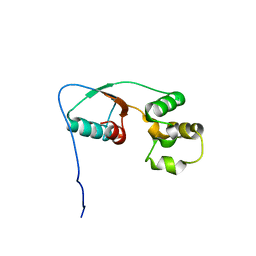 | |
2MU1
 
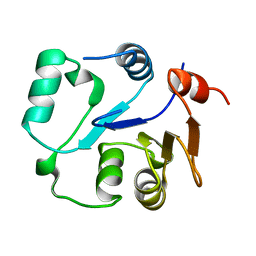 | | NMR structure of the core domain of NP_346487.1, a putative phosphoglycolate phosphatase from Streptococcus pneumoniae TIGR4 | | Descriptor: | Hydrolase, haloacid dehalogenase-like family | | Authors: | Jaudzems, K, Serrano, P, Pedrini, B, Geralt, M, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2014-09-03 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | J-UNIO protocol used for NMR structure determination of the 206-residue protein NP_346487.1 from Streptococcus pneumoniae TIGR4.
J.Biomol.Nmr, 61, 2015
|
|
2MU2
 
 | | NMR structure of the cap domain of NP_346487.1, a putative phosphoglycolate phosphatase from Streptococcus pneumoniae TIGR4 | | Descriptor: | Hydrolase, haloacid dehalogenase-like family | | Authors: | Jaudzems, K, Serrano, P, Pedrini, B, Geralt, M, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2014-09-03 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | J-UNIO protocol used for NMR structure determination of the 206-residue protein NP_346487.1 from Streptococcus pneumoniae TIGR4.
J.Biomol.Nmr, 61, 2015
|
|
2MU3
 
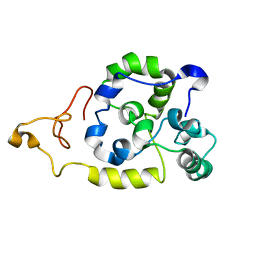 | | Spider wrapping silk fibre architecture arising from its modular soluble protein precursor | | Descriptor: | Aciniform spidroin 1 | | Authors: | Xu, L, Tremblay, M, Meng, Q, Liu, X, Rainey, J.K, Lefevre, T, Sarker, M, Orrell, K.E, Leclerc, J, Pezolet, M, Auger, M. | | Deposit date: | 2014-09-03 | | Release date: | 2015-07-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Spider wrapping silk fibre architecture arising from its modular soluble protein precursor.
Sci Rep, 5, 2015
|
|
2MU4
 
 | | Structure of F. tularensis Virulence Determinant | | Descriptor: | flpp3Sol_2 | | Authors: | Zook, J.J.D.Z, Mo, G.G.M, Craciunescu, F.F.C, Sisco, N.N.S, Hansen, D.D.H, Baravati, B.B.B, Van Horn, W.W.V.H, Cherry, B.B.C, Fromme, P.P.F. | | Deposit date: | 2014-09-03 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Francisella tularensis Virulence Determinant Reveals Structural Homology to Bet v1 Allergen Proteins.
Structure, 23, 2015
|
|
2MU6
 
 | |
2MU7
 
 | | Shortening and modifying the 1513 MSP-1 peptide's alpha-helical region induces protection against malaria | | Descriptor: | 1513 MSP-1 peptide | | Authors: | Espejo, F, Bermudez, A, Torres, E, Urquiza, M, Rodriguez, R, Lopez, Y, Patarroyo, M. | | Deposit date: | 2014-09-04 | | Release date: | 2014-11-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Shortening and modifying the 1513 MSP-1 peptide's alpha-helical region induces protection against malaria.
Biochem.Biophys.Res.Commun., 315, 2004
|
|
2MU8
 
 | | Residues belonging the n-terminal region derived of merozoite surface protein-2 of plasmodium falciparum | | Descriptor: | MSP-2 peptide | | Authors: | Cifuentes, G, Patarroyo, M.E, Urquiza, M, Ramirez, L.E, Reyes, C, Rodriguez, R. | | Deposit date: | 2014-09-04 | | Release date: | 2014-11-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Distorting malaria peptide backbone structure to enable fitting into MHC class II molecules renders modified peptides immunogenic and protective.
J.Med.Chem., 46, 2003
|
|
2MU9
 
 | | Changing ABRA protein peptide to fit the HLA-DR B1*0301 molecule renders it protection-inducing | | Descriptor: | P101/acidic basic repeat antigen | | Authors: | Salazar, L, Alba, M, Curtidor, H, Bermudez, A, Vargas, L, Rivera, Z, Patarroyo, M. | | Deposit date: | 2014-09-04 | | Release date: | 2015-09-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Changing ABRA protein peptide to fit into the HLA-DRbeta1*0301 molecule renders it protection-inducing.
Biochem.Biophys.Res.Commun., 322, 2004
|
|
2MUA
 
 | |
