2M6E
 
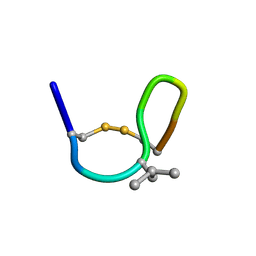 | | NMR solution structure of cis (minor) form of In936 in Methanol | | Descriptor: | Contryphan-In | | Authors: | Sonti, R. | | Deposit date: | 2013-03-29 | | Release date: | 2013-10-30 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Conformational diversity in contryphans from Conus venom: cis-trans isomerisation and aromatic/proline interactions in the 23-membered ring of a 7-residue peptide disulfide loop.
Chemistry, 19, 2013
|
|
2M6F
 
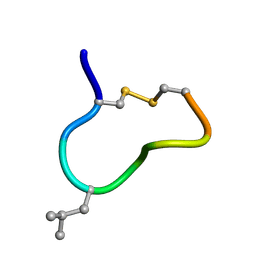 | | NMR solution structure of trans (major) form of In936 in Methanol | | Descriptor: | Contryphan-In | | Authors: | Sonti, R. | | Deposit date: | 2013-03-29 | | Release date: | 2013-10-30 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Conformational diversity in contryphans from Conus venom: cis-trans isomerisation and aromatic/proline interactions in the 23-membered ring of a 7-residue peptide disulfide loop.
Chemistry, 19, 2013
|
|
2M6G
 
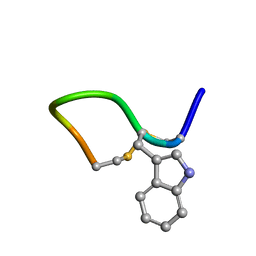 | | Solution structure of cis(C2-P3) trans (D5-P6) form of lO959 in water | | Descriptor: | Contryphan-Lo | | Authors: | Sonti, R. | | Deposit date: | 2013-03-29 | | Release date: | 2013-10-30 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Conformational diversity in contryphans from Conus venom: cis-trans isomerisation and aromatic/proline interactions in the 23-membered ring of a 7-residue peptide disulfide loop.
Chemistry, 19, 2013
|
|
2M6H
 
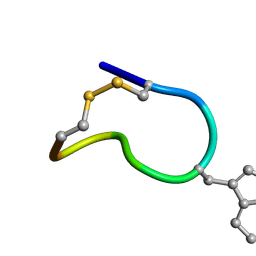 | | Solution structure of trans(C2-P3) trans (D5-P6) of LO959 in methanol | | Descriptor: | Contryphan-Lo | | Authors: | Sonti, R. | | Deposit date: | 2013-03-29 | | Release date: | 2013-10-30 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Conformational diversity in contryphans from Conus venom: cis-trans isomerisation and aromatic/proline interactions in the 23-membered ring of a 7-residue peptide disulfide loop.
Chemistry, 19, 2013
|
|
2M6I
 
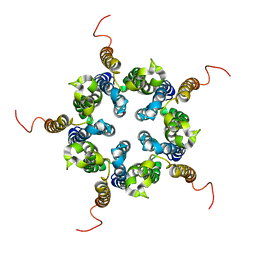 | | Putative pentameric open-channel structure of full-length transmembrane domains of human glycine receptor alpha1 subunit | | Descriptor: | Full-Length Transmembrane Domains of Human Glycine Receptor alpha1 Subunit | | Authors: | Mowrey, D, Cui, T, Jia, Y, Ma, D, Makhov, A.M, Zhang, P, Tang, P, Xu, Y. | | Deposit date: | 2013-03-29 | | Release date: | 2013-09-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Open-Channel Structures of the Human Glycine Receptor alpha 1 Full-Length Transmembrane Domain.
Structure, 21, 2013
|
|
2M6J
 
 | |
2M6K
 
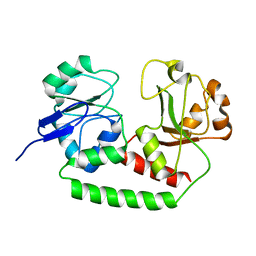 | | Solution structure of the Escherichia coli apo ferric enterobactin binding protein | | Descriptor: | Ferrienterobactin-binding periplasmic protein | | Authors: | Chu, B.C.H, Otten, R, Krewulak, K.D, Mulder, F.A.A, Vogel, H.J. | | Deposit date: | 2013-04-05 | | Release date: | 2014-04-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure, binding properties, and dynamics of the bacterial siderophore-binding protein FepB.
J.Biol.Chem., 289, 2014
|
|
2M6L
 
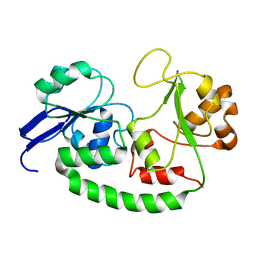 | | Solution structure of the Escherichia coli holo ferric enterobactin binding protein | | Descriptor: | Ferrienterobactin-binding periplasmic protein | | Authors: | Chu, B.C.H, Otten, R, Krewulak, K.D, Mulder, F.A.A, Vogel, H.J. | | Deposit date: | 2013-04-05 | | Release date: | 2014-04-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure, binding properties, and dynamics of the bacterial siderophore-binding protein FepB.
J.Biol.Chem., 289, 2014
|
|
2M6M
 
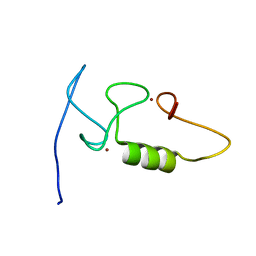 | |
2M6N
 
 | | 3D solution structure of EMI1 (Early Mitotic Inhibitor 1) | | Descriptor: | F-box only protein 5, ZINC ION | | Authors: | Frye, J.J, Brown, N.G, Petzold, G, Watson, E.R, Royappa, G.R, Nourse, A, Jarvis, M, Kriwacki, R.W, Peters, J, Stark, H, Schulman, B.A. | | Deposit date: | 2013-04-06 | | Release date: | 2013-05-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Electron microscopy structure of human APC/C(CDH1)-EMI1 reveals multimodal mechanism of E3 ligase shutdown.
Nat.Struct.Mol.Biol., 20, 2013
|
|
2M6O
 
 | | The actinobacterial transcription factor RbpA binds to the principal sigma subunit of RNA polymerase | | Descriptor: | Uncharacterized protein | | Authors: | Liu, B, Tabib-Salazar, A, Doughty, P, Lewis, R, Ghosh, S, Parsy, M, Simpson, P, Matthews, S, Paget, M. | | Deposit date: | 2013-04-06 | | Release date: | 2013-05-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The actinobacterial transcription factor RbpA binds to the principal sigma subunit of RNA polymerase.
Nucleic Acids Res., 41, 2013
|
|
2M6P
 
 | |
2M6Q
 
 | | Refined Solution NMR Structure of Staphylococcus aureus protein SAV1430. Northeast Strucutral Genomics Consortium Target ZR18 | | Descriptor: | SAV1430 | | Authors: | Baran, M.C, Aramini, J.M, Huang, Y.J, Xiao, R, Acton, T.B, Shih, L, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-04-08 | | Release date: | 2013-05-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | PDBStat: a universal restraint converter and restraint analysis software package for protein NMR.
J.Biomol.Nmr, 56, 2013
|
|
2M6R
 
 | | apo_YqcA | | Descriptor: | Flavodoxin | | Authors: | Jin, C, Hu, Y, Ye, Q. | | Deposit date: | 2013-04-09 | | Release date: | 2014-04-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C and 15N resonance assignments of the apo and holo states of flavodoxin YqcA from Escherichia coli.
Biomol.Nmr Assign., 8, 2014
|
|
2M6S
 
 | | Holo_YqcA | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin | | Authors: | Jin, C, Hu, Y, Ye, Q. | | Deposit date: | 2013-04-09 | | Release date: | 2014-04-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C and 15N resonance assignments of the apo and holo states of flavodoxin YqcA from Escherichia coli.
Biomol.Nmr Assign., 8, 2014
|
|
2M6T
 
 | | NMR solution structure ensemble of 3-4D mutant domain 11 IGF2R | | Descriptor: | Insulin-like growth factor 2 receptor variant | | Authors: | Strickland, M, Williams, C, Richards, E, Minnall, L, Crump, M.P, Frago, S, Hughes, J, Garner, L, Hoppe, H, Rezgui, D, Zaccheo, O.J, Prince, S.N, Hassan, A.B, Whittaker, S. | | Deposit date: | 2013-04-09 | | Release date: | 2014-10-15 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Functional evolution of IGF2:IGF2R domain 11 binding generates novel structural interactions and a specific IGF2 antagonist.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2M6U
 
 | | NMR Structure of CbpAN from Streptococcus pneumoniae | | Descriptor: | Choline binding protein A | | Authors: | Liu, A, Yan, H, Achila, D, Martinez-Hackert, E, Li, Y, Banerjee, R. | | Deposit date: | 2013-04-10 | | Release date: | 2014-04-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural determinants of host specificity of complement Factor H recruitment by Streptococcus pneumoniae.
Biochem.J., 465, 2015
|
|
2M6V
 
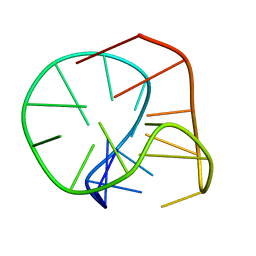 | |
2M6W
 
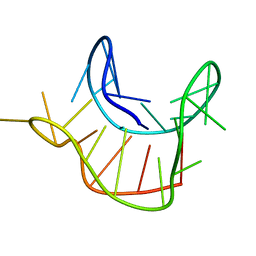 | |
2M6X
 
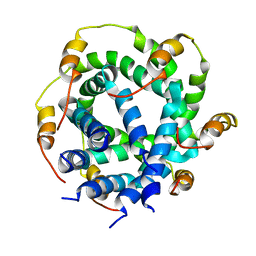 | |
2M6Y
 
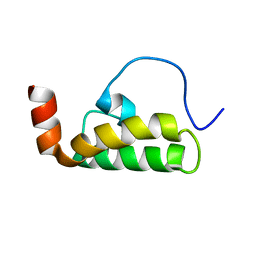 | | The solution structure of the J-domain of human DnaJA1 | | Descriptor: | DnaJ homolog subfamily A member 1 | | Authors: | Stark, J.L, Mehla, K, Chaika, N, Acton, T.B, Xiao, R, Singh, P.K, Montelione, G.T, Powers, R, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-04-14 | | Release date: | 2013-06-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and function of human DnaJ homologue subfamily a member 1 (DNAJA1) and its relationship to pancreatic cancer.
Biochemistry, 53, 2014
|
|
2M6Z
 
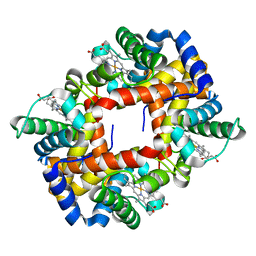 | |
2M70
 
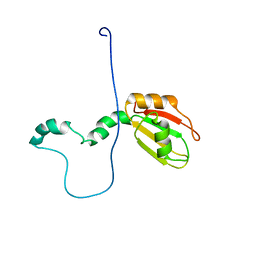 | |
2M71
 
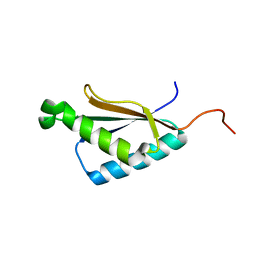 | | Solution structure of the a C-terminal domain of translation initiation factor IF-3 from Campylobacter jejuni | | Descriptor: | Translation initiation factor IF-3 | | Authors: | Harris, R, Ahmed, M, Attonito, J, Bonanno, J.B, Chamala, S, Chowdhury, S, Evans, B, Fiser, A, Glenn, A.S, Hammonds, J, Hillerich, B, Khafizov, K, Lafleur, J, Love, J.D, Seidel, R.D, Stead, M, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-16 | | Release date: | 2013-05-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the a C-terminal domain of translation initiation factor IF-3 from Campylobacter jejuni
To be Published
|
|
2M72
 
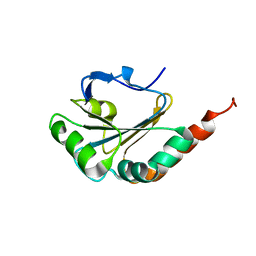 | | Solution structure of uncharacterized thioredoxin-like protein PG_2175 from Porphyromonas gingivalis | | Descriptor: | Uncharacterized thioredoxin-like protein | | Authors: | Harris, R, Ahmed, M, Attonito, J, Bonanno, J.B, Chamala, S, Chowdhury, S, Evans, B, Fiser, A, Glenn, A.S, Hammonds, J, Hillerich, B, Khafizov, K, Lafleur, J, Love, J.D, Seidel, R.D, Stead, M, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-16 | | Release date: | 2013-05-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of uncharacterized thioredoxin-like protein PG_2175 from Porphyromonas gingivalis
To be Published
|
|
