8BOO
 
 | |
8BQY
 
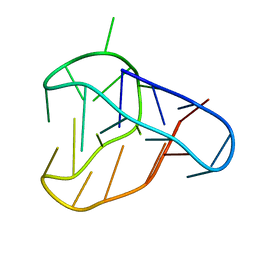 | | An i-motif domain able to undergo pH-dependent conformational transitions (acidic structure) | | Descriptor: | DNA (5'-D(*CP*(DNR)P*GP*TP*TP*(DNR)P*(DNR)P*GP*TP*TP*TP*TP*TP*CP*CP*GP*TP*TP*(DNR)P*CP*GP*T)-3') | | Authors: | Serrano-Chacon, I, Mir, B, Cupellini, L, Colizzi, F, Orozco, M, Escaja, N, Gonzalez, C. | | Deposit date: | 2022-11-22 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | pH-Dependent Capping Interactions Induce Large-Scale Structural Transitions in i-Motifs.
J.Am.Chem.Soc., 145, 2023
|
|
8BSS
 
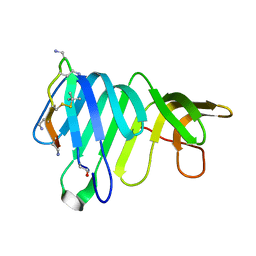 | | Solution Structure of thanatin-like derivative 5 in complex with E. coli LptA mutant Q62L | | Descriptor: | Lipopolysaccharide export system protein LptA, Thanatin-like derivative | | Authors: | Oi, K.K, Jurt, S, Moehle, K, Zerbe, O. | | Deposit date: | 2022-11-26 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Peptidomimetic antibiotics disrupt the lipopolysaccharide transport bridge of drug-resistant Enterobacteriaceae.
Sci Adv, 9, 2023
|
|
8BV6
 
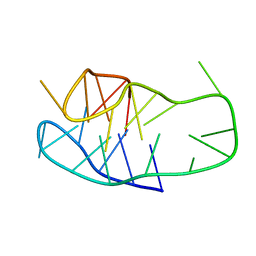 | | An i-motif domain able to undergo pH-dependent conformational transitions (neutral structure) | | Descriptor: | DNA (5'-D(*CP*(DNR)P*GP*TP*TP*CP*(DNR)P*GP*TP*TP*TP*TP*TP*CP*CP*GP*TP*TP*CP*CP*GP*T)-3') | | Authors: | Serrano-Chacon, I, Mir, B, Cupellini, L, Colizzi, F, Orozco, M, Escaja, N, Gonzalez, C. | | Deposit date: | 2022-12-01 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | pH-Dependent Capping Interactions Induce Large-Scale Structural Transitions in i-Motifs.
J.Am.Chem.Soc., 145, 2023
|
|
8BVC
 
 | |
8BVZ
 
 | |
8BWB
 
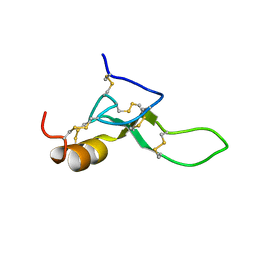 | | Spider toxin Pha1b (PnTx3-6) from Phoneutria nigriventer targeting CaV2.x calcium channels and TRPA1 channel | | Descriptor: | Omega-ctenitoxin-Pn4a | | Authors: | Mironov, P.A, Chernaya, E.M, Paramonov, A.S, Shenkarev, Z.O. | | Deposit date: | 2022-12-06 | | Release date: | 2023-06-21 | | Last modified: | 2023-07-05 | | Method: | SOLUTION NMR | | Cite: | Recombinant Production, NMR Solution Structure, and Membrane Interaction of the Ph alpha 1 beta Toxin, a TRPA1 Modulator from the Brazilian Armed Spider Phoneutria nigriventer .
Toxins, 15, 2023
|
|
8BWT
 
 | |
8BWW
 
 | |
8BXJ
 
 | |
8BZU
 
 | |
8C18
 
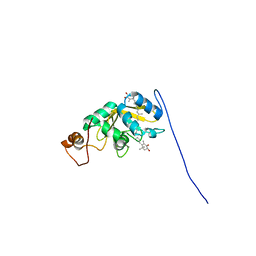 | | Solution structure of carotenoid-binding protein AstaPo1 in complex with astaxanthin | | Descriptor: | ASTAXANTHIN, Astaxanthin binding fasciclin family protein | | Authors: | Kornilov, F.D, Savitskaya, A.G, Slonimskiy, Y.B, Goncharuk, S.A, Sluchanko, N.N, Mineev, K.S. | | Deposit date: | 2022-12-20 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the ligand promiscuity of the neofunctionalized, carotenoid-binding fasciclin domain protein AstaP.
Commun Biol, 6, 2023
|
|
8C2Q
 
 | | Silver ion-bound structure of the silver specific chaperone SilF needed for bacterial silver resistance | | Descriptor: | Copper ABC transporter substrate-binding protein, SILVER ION | | Authors: | Monneau, Y.R, Walker, O, Hologne, M. | | Deposit date: | 2022-12-22 | | Release date: | 2023-10-25 | | Method: | SOLUTION NMR | | Cite: | The battle for silver binding: How the interplay between the SilE, SilF, and SilB proteins contributes to the silver efflux pump mechanism.
J.Biol.Chem., 299, 2023
|
|
8C5J
 
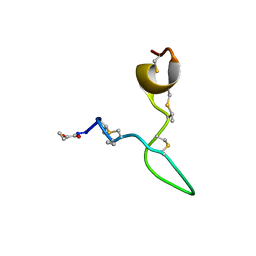 | | Spatial structure of Lch-alpha peptide from two-component lantibiotic system Lichenicidin VK21 | | Descriptor: | Lantibiotic lichenicidin VK21 A1 | | Authors: | Mineev, K.S, Paramonov, A.S, Arseniev, A.S, Ovchinnikova, T.V, Shenkarev, Z.O. | | Deposit date: | 2023-01-09 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Specific Binding of the alpha-Component of the Lantibiotic Lichenicidin to the Peptidoglycan Precursor Lipid II Predetermines Its Antimicrobial Activity.
Int J Mol Sci, 24, 2023
|
|
8C7A
 
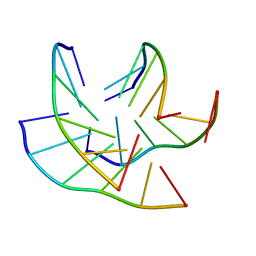 | |
8C7B
 
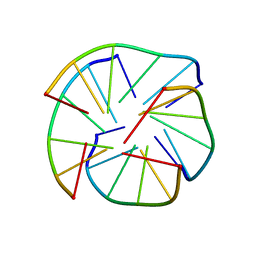 | |
8C7K
 
 | |
8C8A
 
 | | The NMR structure of the MAX28 effector from Magnaporthe oryzae | | Descriptor: | But2 domain-containing protein | | Authors: | Lahfa, M, Padilla, A, de Guillen, K, Kroj, T, Roumestand, C, Barthe, P. | | Deposit date: | 2023-01-19 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | SOLUTION NMR | | Cite: | The structural landscape and diversity of Pyricularia oryzae MAX effectors revisited.
Plos Pathog., 20, 2024
|
|
8CA0
 
 | |
8CF2
 
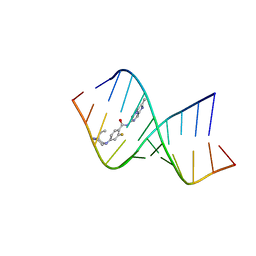 | | Solution structure of the RNA helix formed by the 5'-end of U1 snRNA and an A-1 bulged 5'-splice site in complex with SMN-CY | | Descriptor: | 4-[(3~{S})-3-ethylpiperazin-1-yl]-2-fluoranyl-~{N}-(2-methylimidazo[1,2-a]pyrazin-6-yl)benzamide, RNA (5'-R(P*AP*UP*AP*CP*(PSU)P*(PSU)P*AP*CP*CP*UP*G)-3'), RNA (5'-R(P*GP*GP*AP*GP*UP*AP*AP*GP*UP*CP*U)-3') | | Authors: | Malard, F, Marquevielle, J, Campagne, S. | | Deposit date: | 2023-02-02 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The diversity of splicing modifiers acting on A-1 bulged 5'-splice sites reveals rules for rational drug design.
Nucleic Acids Res., 52, 2024
|
|
8CF4
 
 | |
8CGF
 
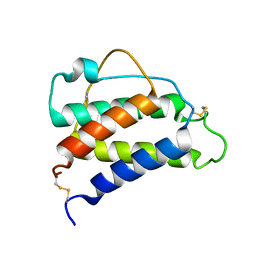 | | Interleukin-4 (wild type) pH 2.4 | | Descriptor: | Interleukin-4 | | Authors: | Vaz, D.C, Rodrigues, J.R, Loureiro-Ferreira, N, Mueller, T, Sebald, W, Redfield, C, Brito, R.M.M. | | Deposit date: | 2023-02-04 | | Release date: | 2023-10-18 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Lessons on protein structure from interleukin-4: All disulfides are not created equal.
Proteins, 92, 2024
|
|
8CH7
 
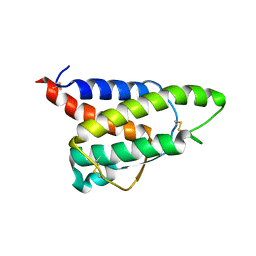 | | RDC-refined Interleukin-4 (wild type) pH 5.6 | | Descriptor: | Interleukin-4 | | Authors: | Vaz, D.C, Rodrigues, J.R, Loureiro-Ferreira, N, Mueller, T, Sebald, W, Redfield, C, Brito, R.M.M. | | Deposit date: | 2023-02-07 | | Release date: | 2023-10-18 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Lessons on protein structure from interleukin-4: All disulfides are not created equal.
Proteins, 92, 2024
|
|
8CHG
 
 | |
8CHH
 
 | |
