6Y8W
 
 | |
6Y94
 
 | | Ca2+-bound Calmodulin mutant N53I | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Holt, C, Nielsen, L.H, Lau, K, Brohus, M, Sorensen, A.B, Larsen, K.T, Sommer, C, Petegem, F.V, Overgaard, M.T, Wimmer, R. | | Deposit date: | 2020-03-06 | | Release date: | 2020-04-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The arrhythmogenic N53I variant subtly changes the structure and dynamics in the calmodulin N-terminal domain, altering its interaction with the cardiac ryanodine receptor.
J.Biol.Chem., 295, 2020
|
|
6Y95
 
 | | Ca2+-free Calmodulin mutant N53I | | Descriptor: | Calmodulin | | Authors: | Holt, C, Hamborg, L.N, Lau, K, Brohus, M, Sorensen, A.B, Larsen, K.T, Sommer, C, Petegem, F.V, Overgaard, M.T, Wimmer, R. | | Deposit date: | 2020-03-06 | | Release date: | 2020-04-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The arrhythmogenic N53I variant subtly changes the structure and dynamics in the calmodulin N-terminal domain, altering its interaction with the cardiac ryanodine receptor.
J.Biol.Chem., 295, 2020
|
|
6Y96
 
 | | solution structure of cold-shock domain 9 of drosophila Upstream of N-Ras (Unr) | | Descriptor: | Upstream of N-ras, isoform A | | Authors: | Sweetapple, L.J, Hollmann, N.M, Simon, B, Hennig, J. | | Deposit date: | 2020-03-06 | | Release date: | 2020-07-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Pseudo-RNA-Binding Domains Mediate RNA Structure Specificity in Upstream of N-Ras.
Cell Rep, 32, 2020
|
|
6YCV
 
 | |
6YDH
 
 | |
6YE5
 
 | | Structure of ribosomal binding factor A RbfA of Staphylococcus aureus bacterium by NMR | | Descriptor: | Ribosome-binding factor A | | Authors: | Blokhin, D.S, Usachev, K.S, Bikmullin, A.G, Nurullina, L, Garaeva, N, Validov, S, Klochkov, V, Aganov, A, Khusainov, I, Yusupov, M. | | Deposit date: | 2020-03-24 | | Release date: | 2021-03-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure of ribosomal binding factor A RbfA of Staphylococcus aureus bacterium by NMR
To Be Published
|
|
6YEG
 
 | | Hybrid structure of the SPP1 tail tube by solid-state NMR and cryo EM - Final EM Refinement | | Descriptor: | Tail tube protein gp17.1* | | Authors: | Zinke, M, Sachowsky, K.A.A, Zinn-Justin, S, Ravelli, R, Schroder, G.F, Habeck, M, Lange, A. | | Deposit date: | 2020-03-24 | | Release date: | 2020-10-14 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (4 Å), SOLID-STATE NMR | | Cite: | Architecture of the flexible tail tube of bacteriophage SPP1.
Nat Commun, 11, 2020
|
|
6YEL
 
 | | Stromal interaction molecule 1 coiled-coil 1 fragment | | Descriptor: | Stromal interaction molecule 1 | | Authors: | Rathner, P, Cerofolini, L, Ravera, E, Bechmann, M, Grabmayr, H, Fahrner, M, Fragai, M, Romanin, C, Luchinat, C, Mueller, N. | | Deposit date: | 2020-03-25 | | Release date: | 2020-09-02 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Interhelical interactions within the STIM1 CC1 domain modulate CRAC channel activation.
Nat.Chem.Biol., 17, 2021
|
|
6YEP
 
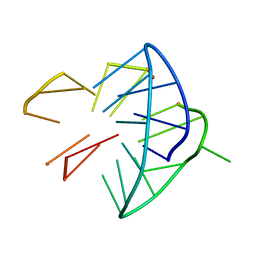 | |
6YET
 
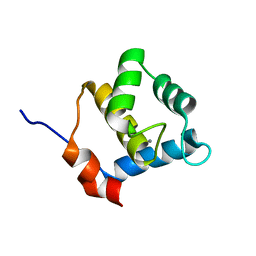 | | Second EH domain of AtEH1/Pan1 | | Descriptor: | CALCIUM ION, Calcium-binding EF hand family protein | | Authors: | Yperman, K, Papageorgiou, A, Evangelidis, T, Van Damme, D, Tripsianes, K. | | Deposit date: | 2020-03-25 | | Release date: | 2021-03-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Distinct EH domains of the endocytic TPLATE complex confer lipid and protein binding.
Nat Commun, 12, 2021
|
|
6YEU
 
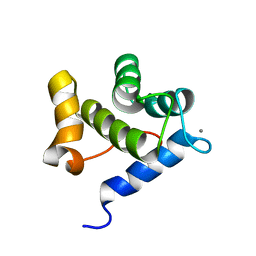 | | Second EH domain of AtEH1/Pan1 | | Descriptor: | CALCIUM ION, Calcium-binding EF hand family protein | | Authors: | Yperman, K, Papageorgiou, A, Evangelidis, T, Van Damme, D, Tripsianes, K. | | Deposit date: | 2020-03-25 | | Release date: | 2021-03-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Distinct EH domains of the endocytic TPLATE complex confer lipid and protein binding.
Nat Commun, 12, 2021
|
|
6YFY
 
 | | Solid-state NMR structure of the D-Arg4,L10-teixobactin - Lipid II complex in lipid bilayers. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-alpha-muramic acid, 3-methylbut-2-en-1-ol, D-Arg4,Leu10-Teixobactin, ... | | Authors: | Weingarth, M.H, Shukla, R. | | Deposit date: | 2020-03-26 | | Release date: | 2020-06-10 | | Last modified: | 2024-08-07 | | Method: | SOLID-STATE NMR | | Cite: | Mode of action of teixobactins in cellular membranes.
Nat Commun, 11, 2020
|
|
6YHF
 
 | | Solution NMR Structure of APP TMD | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Silber, M, Muhle-Goll, C. | | Deposit date: | 2020-03-29 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Altered Hinge Conformations in APP Transmembrane Helix Mutants May Affect Enzyme-Substrate Interactions of gamma-Secretase.
Acs Chem Neurosci, 11, 2020
|
|
6YHI
 
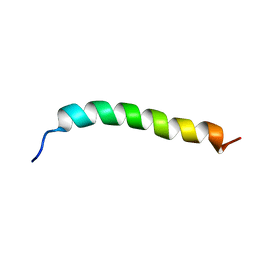 | | Solution NMR Structure of APP G38L mutant TMD | | Descriptor: | Amyloid-beta precursor protein G38L mutant | | Authors: | Silber, M, Muhle-Goll, C. | | Deposit date: | 2020-03-30 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Altered Hinge Conformations in APP Transmembrane Helix Mutants May Affect Enzyme-Substrate Interactions of gamma-Secretase.
Acs Chem Neurosci, 11, 2020
|
|
6YHO
 
 | | Solution NMR Structure of APP G38P mutant TM | | Descriptor: | Amyloid-beta precursor protein G38P mutant | | Authors: | Silber, M, Muhle-Goll, C. | | Deposit date: | 2020-03-30 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Altered Hinge Conformations in APP Transmembrane Helix Mutants May Affect Enzyme-Substrate Interactions of gamma-Secretase.
Acs Chem Neurosci, 11, 2020
|
|
6YHP
 
 | | Solution NMR Structure of APP V44M mutant TMD | | Descriptor: | Amyloid-beta precursor protein V44M mutant | | Authors: | Silber, M, Muhle-Goll, C. | | Deposit date: | 2020-03-30 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Altered Hinge Conformations in APP Transmembrane Helix Mutants May Affect Enzyme-Substrate Interactions of gamma-Secretase.
Acs Chem Neurosci, 11, 2020
|
|
6YHX
 
 | | Solution NMR Structure of APP I45T mutant TMD | | Descriptor: | Amyloid-beta precursor protein I45T mutant | | Authors: | Silber, M, Muhle-Goll, C. | | Deposit date: | 2020-03-31 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Altered Hinge Conformations in APP Transmembrane Helix Mutants May Affect Enzyme-Substrate Interactions of gamma-Secretase.
Acs Chem Neurosci, 11, 2020
|
|
6YHZ
 
 | |
6YI2
 
 | |
6YI3
 
 | |
6YJ0
 
 | |
6YJL
 
 | |
6YP5
 
 | |
6YQ5
 
 | | Hybrid structure of the SPP1 tail tube by solid-state NMR and cryo EM - NMR Ensemble | | Descriptor: | Tail tube protein gp17.1* | | Authors: | Zinke, M, Sachowsky, K.A.A, Zinn-Justin, S, Ravelli, R, Schroder, G.F, Habeck, M, Lange, A. | | Deposit date: | 2020-04-16 | | Release date: | 2020-10-14 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4 Å), SOLID-STATE NMR | | Cite: | Architecture of the flexible tail tube of bacteriophage SPP1.
Nat Commun, 11, 2020
|
|
