6R9Z
 
 | |
6RC7
 
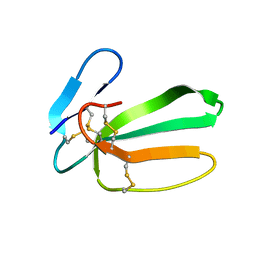 | | NMR structure of cytotoxin 3 from Naja kaouthia in solution, major form | | Descriptor: | Cytotoxin 3 | | Authors: | Dubinnyi, M.A, Dubovskii, P.V, Utkin, Y.N, Arseniev, A.S. | | Deposit date: | 2019-04-11 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | The omega-loop of cobra cytotoxins tolerates multiple amino acid substitutions.
Biochem.Biophys.Res.Commun., 558, 2021
|
|
6RFM
 
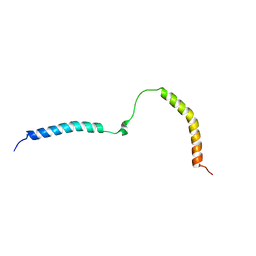 | |
6RH5
 
 | | Solution structure and 1H, 13C and 15N chemical shift assignments for NECAP1 PHear domain | | Descriptor: | Adaptin ear-binding coat-associated protein 1 | | Authors: | Owen, D.J, Neuhaus, D, Yang, J.-C, Herrmann, T. | | Deposit date: | 2019-04-18 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Temporal Ordering in Endocytic Clathrin-Coated Vesicle Formation via AP2 Phosphorylation.
Dev.Cell, 50, 2019
|
|
6RH6
 
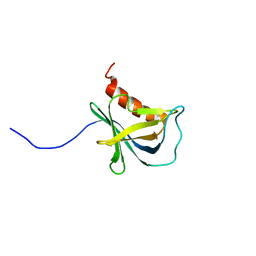
 | | Solution structure and 1H, 13C and 15N chemical shift assignments for the complex of NECAP1 PHear domain with phosphorylated AP2 mu2 148-163 | | Descriptor: | AP-2 complex subunit mu, Adaptin ear-binding coat-associated protein 1 | | Authors: | Owen, D.J, Neuhaus, D, Yang, J.-C, Herrmann, T. | | Deposit date: | 2019-04-18 | | Release date: | 2019-09-04 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Temporal Ordering in Endocytic Clathrin-Coated Vesicle Formation via AP2 Phosphorylation.
Dev.Cell, 50, 2019
|
|
6RHY
 
 | |
6RIO
 
 | | Imidazole Polyamide-DNA complex NMR structure (5'-CGATGTACATCG-3') | | Descriptor: | 3-[3-[[4-[[4-[[4-[[4-[[(2~{R})-2-azaniumyl-4-[[1-methyl-4-[[1-methyl-4-[[1-methyl-4-[(1-methylimidazol-2-yl)carbonylamino]pyrrol-2-yl]carbonylamino]pyrrol-2-yl]carbonylamino]pyrrol-2-yl]carbonylamino]butanoyl]amino]-1-methyl-imidazol-2-yl]carbonylamino]-1-methyl-pyrrol-2-yl]carbonylamino]-1-methyl-pyrrol-2-yl]carbonylamino]-1-methyl-pyrrol-2-yl]carbonylamino]propanoylamino]propyl-dimethyl-azanium, DNA (5'-(*(DC5)P*GP*AP*TP*GP*TP*AP*CP*AP*TP*CP*(DG3))-3') | | Authors: | Padroni, G, Withers, J.M, Taladriz-Sender, A, Reichenbach, L.F, Parkinson, J.A, Burley, G.A. | | Deposit date: | 2019-04-24 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Sequence-Selective Minor Groove Recognition of a DNA Duplex Containing Synthetic Genetic Components.
J.Am.Chem.Soc., 141, 2019
|
|
6RK3
 
 | | Solution structure of the ribosome Elongation Factor P (EF-P) from Staphylococcus aureus | | Descriptor: | Elongation factor P | | Authors: | Usachev, K, Fatkhullin, B, Gabdulkhakov, A, Khusainov, I, Golubev, A, Validov, S, Yusupova, G, Yusupov, M. | | Deposit date: | 2019-04-30 | | Release date: | 2020-03-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR and crystallographic structural studies of the Elongation factor P from Staphylococcus aureus.
Eur.Biophys.J., 49, 2020
|
|
6RLS
 
 | | Concerted dynamics of metallo-base pairs in an A/B-form helical transition (apo species) | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*TP*CP*AP*TP*GP*AP*TP*AP*CP*G)-3')_apo | | Authors: | Schmidt, O.P, Jurt, S, Johannsen, S, Karimi, A, Sigel, R.K.O, Luedtke, N.W. | | Deposit date: | 2019-05-02 | | Release date: | 2019-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Concerted dynamics of metallo-base pairs in an A/B-form helical transition.
Nat Commun, 10, 2019
|
|
6RPV
 
 | | Extremely stable monomeric variant of human cystatin C with single amino acid substitution | | Descriptor: | Cystatin-C | | Authors: | Zhukov, I, Rodziewicz-Motowidlo, S, Maszota-Zieleniak, M, Jurczak, P, Kozak, M. | | Deposit date: | 2019-05-14 | | Release date: | 2019-07-31 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | NMR and crystallographic structural studies of the extremely stable monomeric variant of human cystatin C with single amino acid substitution.
Febs J., 287, 2020
|
|
6RQS
 
 | | RW16 peptide | | Descriptor: | ARG-ARG-TRP-ARG-ARG-TRP-TRP-ARG-ARG-TRP-TRP-ARG-ARG-TRP-ARG-ARG | | Authors: | Jobin, M.-L, Grelard, A, Alves, I, Mackereth, C.D. | | Deposit date: | 2019-05-16 | | Release date: | 2019-10-23 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Biophysical Insight on the Membrane Insertion of an Arginine-Rich Cell-Penetrating Peptide.
Int J Mol Sci, 20, 2019
|
|
6RRL
 
 | |
6RRO
 
 | |
6RS3
 
 | |
6RSF
 
 | | NMR structure of pleurocidin KR in SDS micelles | | Descriptor: | Pleurocidin | | Authors: | Manzo, G, Mason, A.J. | | Deposit date: | 2019-05-21 | | Release date: | 2020-12-09 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A pleurocidin analogue with greater conformational flexibility, enhanced antimicrobial potency and in vivo therapeutic efficacy.
Commun Biol, 3, 2020
|
|
6RSG
 
 | | NMR structure of pleurocidin VA in SDS micelles | | Descriptor: | Pleurocidin | | Authors: | Manzo, G, Mason, A.J. | | Deposit date: | 2019-05-21 | | Release date: | 2020-12-09 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A pleurocidin analogue with greater conformational flexibility, enhanced antimicrobial potency and in vivo therapeutic efficacy.
Commun Biol, 3, 2020
|
|
6RSM
 
 | | Solution NMR structure of the peptide 12530 from medicinal leech Hirudo medicinalis in dodecylphosphocholine micelles | | Descriptor: | peptide 12530 | | Authors: | Talyzina, I.A, Nadezhdin, K.D, Grafskaia, E.N, Arseniev, A.S, Lazarev, V.N. | | Deposit date: | 2019-05-21 | | Release date: | 2019-07-24 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Medicinal leech antimicrobial peptides lacking toxicity represent a promising alternative strategy to combat antibiotic-resistant pathogens.
Eur.J.Med.Chem., 180, 2019
|
|
6RSS
 
 | | Solution structure of the fourth WW domain of WWP2 with GB1-tag | | Descriptor: | NEDD4-like E3 ubiquitin-protein ligase WWP2 | | Authors: | Wahl, L.C, Watt, J.E, Tolchard, J, Blumenschein, T.M.A, Chantry, A. | | Deposit date: | 2019-05-22 | | Release date: | 2019-10-09 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Smad7 Binds Differently to Individual and Tandem WW3 and WW4 Domains of WWP2 Ubiquitin Ligase Isoforms.
Int J Mol Sci, 20, 2019
|
|
6RVA
 
 | | STRUCTURE OF [ASP58]-IGF-I ANALOGUE | | Descriptor: | Insulin-like growth factor I | | Authors: | Jiracek, J, Zakova, L, Socha, O. | | Deposit date: | 2019-05-31 | | Release date: | 2019-10-02 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Mutations at hypothetical binding site 2 in insulin and insulin-like growth factors 1 and 2 result in receptor- and hormone-specific responses.
J.Biol.Chem., 294, 2019
|
|
6RWG
 
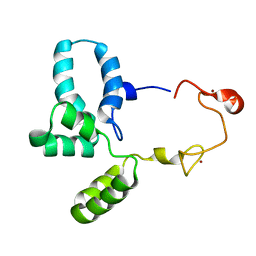 | | Structure of HIV-1 CAcSP1NC mutant(W41A,M42A) interacting with maturation inhibitor EP39 | | Descriptor: | Gag polyprotein, ZINC ION | | Authors: | Chen, X, Coric, P, Larue, V, Nonin-Lecomte, S, Bouaziz, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-05 | | Release date: | 2020-05-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The HIV-1 maturation inhibitor, EP39, interferes with the dynamic helix-coil equilibrium of the CA-SP1 junction of Gag.
Eur.J.Med.Chem., 204, 2020
|
|
6RY9
 
 | |
6RYQ
 
 | |
6RZ1
 
 | |
6RZC
 
 | |
6S0N
 
 | | A9 peptide derived from Herceptin fab binding region | | Descriptor: | GLN-ASP-VAL-ASN-THR-ALA-VAL-ALA-TRP | | Authors: | De Luca, S, Verdoliva, V, Saviano, M, Fattorusso, R, Diana, D. | | Deposit date: | 2019-06-17 | | Release date: | 2019-11-06 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | SPR and NMR characterization of the molecular interaction between A9 peptide and a model system of HER2 receptor: A fragment approach for selecting peptide structures specific for their target.
J.Pept.Sci., 26, 2020
|
|
