2K9X
 
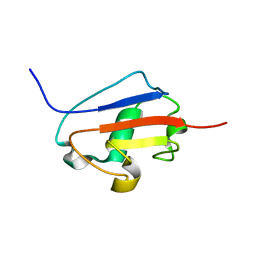 | | Solution structure of Urm1 from Trypanosoma brucei | | Descriptor: | Uncharacterized protein | | Authors: | Zhang, W, Zhang, J, Xu, C, Wang, T, Zhang, X, Tu, X. | | Deposit date: | 2008-10-27 | | Release date: | 2009-03-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Urm1 from Trypanosoma brucei
Proteins, 75, 2009
|
|
2K9Y
 
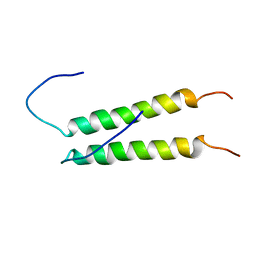 | |
2K9Z
 
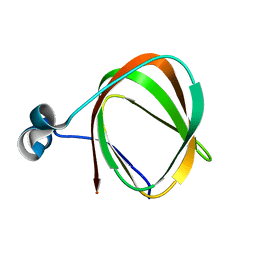 | | NMR structure of the protein TM1112 | | Descriptor: | uncharacterized protein TM1112 | | Authors: | Mohanty, B, Pedrini, B, Serrano, P, Geralt, M, Horst, R, Herrmann, T, Wilson, I.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-10-28 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Comparison of NMR and crystal structures for the proteins TM1112 and TM1367.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2KA0
 
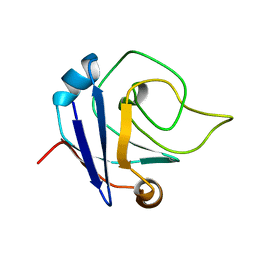 | | NMR structure of the protein TM1367 | | Descriptor: | uncharacterized protein TM1367 | | Authors: | Mohanty, B, Pedrini, B, Serrano, P, Geralt, M, Horst, R, Herrmann, T, Wilson, I.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-10-27 | | Release date: | 2009-01-13 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Comparison of NMR and crystal structures for the proteins TM1112 and TM1367.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2KA1
 
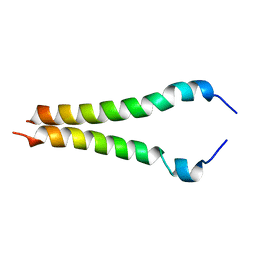 | |
2KA2
 
 | |
2KA3
 
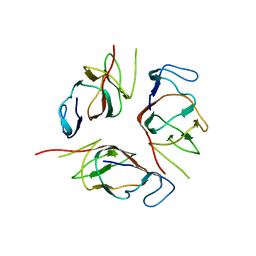 | | Structure of EMILIN-1 C1Q-like domain | | Descriptor: | EMILIN-1 | | Authors: | Verdone, G, Corazza, A, Colebrooke, S.A, Cicero, D.O, Eliseo, T, Boyd, J, Doliana, R, Fogolari, F, Viglino, P, Colombatti, A, Campbell, I.D, Esposito, G. | | Deposit date: | 2008-10-30 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR-based homology model for the solution structure of the C-terminal globular domain of EMILIN1
J.Biomol.Nmr, 43, 2009
|
|
2KA4
 
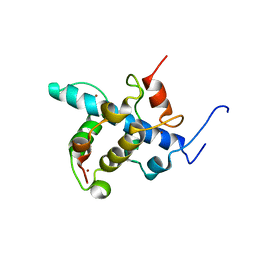 | | NMR structure of the CBP-TAZ1/STAT2-TAD complex | | Descriptor: | Crebbp protein, Signal transducer and activator of transcription 2, ZINC ION | | Authors: | Wojciak, J.M, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2008-10-30 | | Release date: | 2009-04-21 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recruitment of CBP/p300 coactivators by STAT1 and STAT2 transactivation domains
Embo J., 28, 2009
|
|
2KA5
 
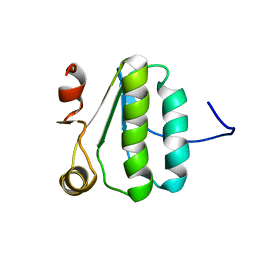 | | NMR Structure of the protein TM1081 | | Descriptor: | Putative anti-sigma factor antagonist TM_1081 | | Authors: | Serrano, P, Geralt, M, Mohanty, B, Pedrini, B, Horst, R, Wuthrich, K, Wilson, I, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-10-30 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Comparison of NMR and crystal structures highlights conformational isomerism in protein active sites.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2KA6
 
 | | NMR structure of the CBP-TAZ2/STAT1-TAD complex | | Descriptor: | CREB-binding protein, Signal transducer and activator of transcription 1-alpha/beta, ZINC ION | | Authors: | Wojciak, J.M, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2008-10-30 | | Release date: | 2009-04-07 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recruitment of CBP/p300 coactivators by STAT1 and STAT2 transactivation domains.
Embo J., 28, 2009
|
|
2KA7
 
 | | NMR solution structure of TM0212 at 40 C | | Descriptor: | Glycine cleavage system H protein | | Authors: | Pedrini, B, Herrmann, T, Mohanty, B, Geralt, M, Wilson, I, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-10-31 | | Release date: | 2009-01-13 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | The J-UNIO protocol for automated protein structure determination by NMR in solution.
J.Biomol.Nmr, 53, 2012
|
|
2KA9
 
 | | Solution structure of PSD-95 PDZ12 complexed with cypin peptide | | Descriptor: | Disks large homolog 4, cypin peptide | | Authors: | Wang, W.N, Weng, J.W, Zhang, X, Liu, M.L, Zhang, M.J. | | Deposit date: | 2008-11-03 | | Release date: | 2009-06-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Creating conformational entropy by increasing interdomain mobility in ligand binding regulation: a revisit to N-terminal tandem PDZ domains of PSD-95
J.Am.Chem.Soc., 131, 2009
|
|
2KAA
 
 | |
2KAC
 
 | |
2KAD
 
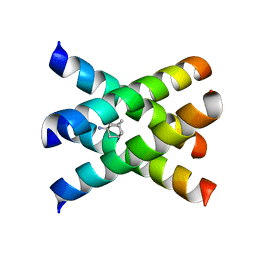 | | Magic-Angle-Spinning Solid-State NMR Structure of Influenza A M2 Transmembrane Domain | | Descriptor: | (3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-amine, Transmembrane peptide of Matrix protein 2 | | Authors: | Hong, M, Cady, S.D, Mishanina, T.V. | | Deposit date: | 2008-11-04 | | Release date: | 2008-11-18 | | Last modified: | 2024-05-22 | | Method: | SOLID-STATE NMR | | Cite: | Structure of amantadine-bound M2 transmembrane peptide of influenza A in lipid bilayers from magic-angle-spinning solid-state NMR: the role of Ser31 in amantadine binding.
J.Mol.Biol., 385, 2009
|
|
2KAE
 
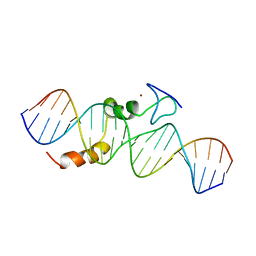 | | data-driven model of MED1:DNA complex | | Descriptor: | 5'-D(*DCP*DGP*DGP*DAP*DAP*DAP*DAP*DGP*DTP*DAP*DTP*DAP*DCP*DTP*DTP*DTP*DTP*DCP*DCP*DG)-3', GATA-type transcription factor, ZINC ION | | Authors: | Lowry, J.A, Gamsjaeger, R, Thong, S, Hung, W, Kwan, A.H, Broitman-Maduro, G, Matthews, J.M, Maduro, M, Mackay, J.P. | | Deposit date: | 2008-11-04 | | Release date: | 2009-01-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of MED-1 Reveals Unexpected Diversity in the Mechanism of DNA Recognition by GATA-type Zinc Finger Domains.
J.Biol.Chem., 284, 2009
|
|
2KAF
 
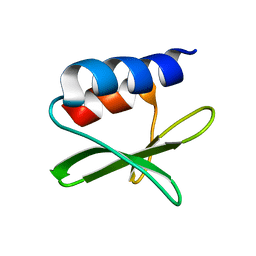 | | Solution structure of the SARS-unique domain-C from the nonstructural protein 3 (nsp3) of the severe acute respiratory syndrome coronavirus | | Descriptor: | Non-structural protein 3 | | Authors: | Johnson, M.A, Mohanty, B, Pedrini, B, Serrano, P, Chatterjee, A, Herrmann, T, Joseph, J, Saikatendu, K, Wilson, I.A, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-11-05 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | SARS coronavirus unique domain: three-domain molecular architecture in solution and RNA binding.
J.Mol.Biol., 400, 2010
|
|
2KAJ
 
 | | NMR structure of gallium substituted ferredoxin | | Descriptor: | Ferredoxin-1, GALLIUM (III) ION | | Authors: | Xu, X, Ubbink, M, Knaff, D.B. | | Deposit date: | 2008-11-06 | | Release date: | 2009-11-10 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural and functional characterization of the ga-substituted ferredoxin from Synechocystis sp. PCC6803, a mimic of the native protein.
Biochemistry, 49, 2010
|
|
2KAK
 
 | | Solution structure of the beta-E-domain of wheat Ec-1 metallothionein | | Descriptor: | EC protein I/II, ZINC ION | | Authors: | Peroza, E.A, Schmucki, R, Guntert, P, Freisinger, E, Zerbe, O. | | Deposit date: | 2008-11-06 | | Release date: | 2009-05-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The beta(E)-domain of wheat E(c)-1 metallothionein: a metal-binding domain with a distinctive structure.
J.Mol.Biol., 387, 2009
|
|
2KAL
 
 | | NMR structure of fully methylated GATC site | | Descriptor: | 5'-D(*DCP*DGP*DCP*DAP*DGP*(6MA)P*DTP*DCP*DTP*DCP*DGP*DC)-3', 5'-D(*DGP*DCP*DGP*DAP*DGP*(6MA)P*DTP*DCP*DTP*DGP*DCP*DG)-3' | | Authors: | Bang, J, Bae, S, Park, C, Lee, J, Choi, B. | | Deposit date: | 2008-11-09 | | Release date: | 2009-02-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamics study of DNA dodecamer duplexes that contain un-, hemi-, or fully methylated GATC sites.
J.Am.Chem.Soc., 130, 2008
|
|
2KAM
 
 | |
2KAN
 
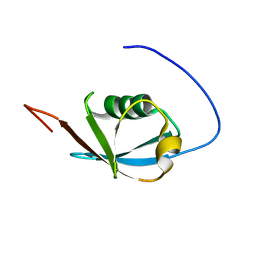 | | Solution NMR structure of ubiquitin-like domain of Arabidopsis thaliana protein At2g32350. Northeast Structural Genomics Consortium target AR3433A | | Descriptor: | uncharacterized protein ar3433a | | Authors: | Eletsky, A, Mills, J.L, Sukumaran, D, Hua, J, Shastry, R, Jiang, M, Ciccosanti, C, Xiao, R, Liu, J, Everret, J.K, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-11-11 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of ubiquitin-like domain of Arabidopsis thaliana protein At2g32350.
To be Published
|
|
2KAP
 
 | | Solution structure of DLC1-SAM | | Descriptor: | Rho GTPase-activating protein 7 | | Authors: | Yang, S, Yang, D. | | Deposit date: | 2008-11-12 | | Release date: | 2009-10-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Characterization of DLC1-SAM equilibrium unfolding at the amino acid residue level
Biochemistry, 48, 2009
|
|
2KAR
 
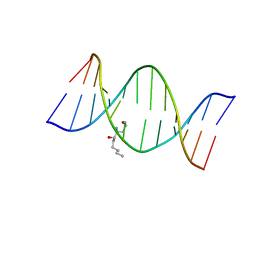 | | HNE-dG adduct mismatched with dA in acidic solution | | Descriptor: | (4S)-nonane-1,4-diol, 5'-D(*DGP*DCP*DTP*DAP*DGP*DCP*DGP*DAP*DGP*DTP*DCP*DC)-3', 5'-D(*DGP*DGP*DAP*DCP*DTP*DAP*DGP*DCP*DTP*DAP*DGP*DC)-3' | | Authors: | Huang, H, Wang, H, Lloyd, R.S, Rizzo, C.J, Stone, M.P. | | Deposit date: | 2008-11-13 | | Release date: | 2009-05-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Conformational interconversion of the trans-4-hydroxynonenal-derived (6S,8R,11S) 1,N(2)-deoxyguanosine adduct when mismatched with deoxyadenosine in DNA
Chem.Res.Toxicol., 22, 2009
|
|
2KAS
 
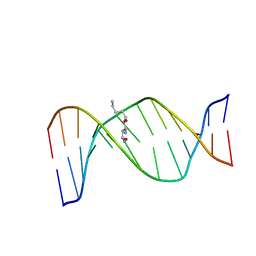 | | HNE-dG adduct mismatched with dA in basic solution | | Descriptor: | (4S)-nonane-1,4-diol, 5'-D(*DGP*DCP*DTP*DAP*DGP*DCP*DGP*DAP*DGP*DTP*DCP*DC)-3', 5'-D(*DGP*DGP*DAP*DCP*DTP*DAP*DGP*DCP*DTP*DAP*DGP*DC)-3' | | Authors: | Huang, H, Wang, H, Lloyd, R.S, Rizzo, C.J, Stone, M.P. | | Deposit date: | 2008-11-13 | | Release date: | 2009-05-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Conformational interconversion of the trans-4-hydroxynonenal-derived (6S,8R,11S) 1,N(2)-deoxyguanosine adduct when mismatched with deoxyadenosine in DNA
Chem.Res.Toxicol., 22, 2009
|
|
