6UCQ
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome recycling complex | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Zhou, D, Tanzawa, T, Gagnon, M.G, Lin, J. | | Deposit date: | 2019-09-17 | | Release date: | 2019-12-25 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for ribosome recycling by RRF and tRNA.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6UDJ
 
 | | HIV-1 bNAb 1-18 in complex with BG505 SOSIP.664 and 10-1074 | | Descriptor: | 1-18 Fab Heavy Chain, 1-18 Fab Light Chain, 10-1074 Fab Heavy Chain, ... | | Authors: | Abernathy, M.E, Barnes, C.O, Gristick, H.B, Bjorkman, P.J. | | Deposit date: | 2019-09-19 | | Release date: | 2020-01-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Restriction of HIV-1 Escape by a Highly Broad and Potent Neutralizing Antibody.
Cell, 180, 2020
|
|
6UE0
 
 | | Crystal structure of dihydrodipicolinate synthase from Klebsiella pneumoniae bound to pyruvate | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate synthase, CHLORIDE ION, SULFATE ION | | Authors: | Impey, R.E, Lee, M, Hawkins, D.A, Sutton, J.M, Panjikar, S, Perugini, M.A, Soares da Costa, T.P. | | Deposit date: | 2019-09-20 | | Release date: | 2020-02-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Mis-annotations of a promising antibiotic target in high-priority gram-negative pathogens.
Febs Lett., 594, 2020
|
|
6UO1
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with mRNA (containing pseudouridine at the first position of the codon) and deacylated A-, P-, and E-site tRNAs at 2.95A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Batool, Z, Dobosz-Bartoszek, M, Polikanov, Y.S. | | Deposit date: | 2019-10-14 | | Release date: | 2019-11-27 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Pseudouridinylation of mRNA coding sequences alters translation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6UTV
 
 | | E. coli sigma-S transcription initiation complex with a 6-nt RNA ("Fresh" crystal soaked with CTP, UTP, GTP, and ddATP for 150 seconds) | | Descriptor: | DIPHOSPHATE, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Zuo, Y, De, S, Steitz, T.A. | | Deposit date: | 2019-10-30 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Structural Insights into Transcription Initiation from De Novo RNA Synthesis to Transitioning into Elongation.
Iscience, 23, 2020
|
|
6UTW
 
 | | E. coli sigma-S transcription initiation complex with a 4-nt RNA ("Fresh" crystal) | | Descriptor: | DIPHOSPHATE, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Zuo, Y, De, S, Steitz, T.A. | | Deposit date: | 2019-10-30 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.854 Å) | | Cite: | Structural Insights into Transcription Initiation from De Novo RNA Synthesis to Transitioning into Elongation.
Iscience, 23, 2020
|
|
6UTX
 
 | | E. coli sigma-S transcription initiation complex with an empty bubble ("Old" crystal) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zuo, Y, De, S, Steitz, T.A. | | Deposit date: | 2019-10-30 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.05 Å) | | Cite: | Structural Insights into Transcription Initiation from De Novo RNA Synthesis to Transitioning into Elongation.
Iscience, 23, 2020
|
|
6UTY
 
 | | E. coli sigma-S transcription initiation complex with a mismatching CTP ("Old" crystal soaked with CTP for 30 minutes) | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Zuo, Y, De, S, Steitz, T.A. | | Deposit date: | 2019-10-30 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.15 Å) | | Cite: | Structural Insights into Transcription Initiation from De Novo RNA Synthesis to Transitioning into Elongation.
Iscience, 23, 2020
|
|
6UTZ
 
 | | E. coli sigma-S transcription initiation complex with a 6-nt RNA ("Fresh" crystal soaked with CTP and UTP for 30 minutes) | | Descriptor: | DIPHOSPHATE, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Zuo, Y, De, S, Steitz, T.A. | | Deposit date: | 2019-10-30 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.803 Å) | | Cite: | Structural Insights into Transcription Initiation from De Novo RNA Synthesis to Transitioning into Elongation.
Iscience, 23, 2020
|
|
6UU0
 
 | | E. coli sigma-S transcription initiation complex with a 3-nt RNA and a mismatching GTP ("Fresh" crystal soaked with GTP for 1 hour) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zuo, Y, De, S, Steitz, T.A. | | Deposit date: | 2019-10-30 | | Release date: | 2020-08-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural Insights into Transcription Initiation from De Novo RNA Synthesis to Transitioning into Elongation.
Iscience, 23, 2020
|
|
6UU1
 
 | | E. coli sigma-S transcription initiation complex with a 4-nt RNA and a CTP ("Fresh" crystal soaked with CTP, GTP, and ddTTP for 30 minutes) | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Zuo, Y, De, S, Steitz, T.A. | | Deposit date: | 2019-10-30 | | Release date: | 2020-08-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (4.097 Å) | | Cite: | Structural Insights into Transcription Initiation from De Novo RNA Synthesis to Transitioning into Elongation.
Iscience, 23, 2020
|
|
6UU2
 
 | | E. coli sigma-S transcription initiation complex with 3-nt RNA ("Old" crystal soaked with GTP and ATP for 30 minutes) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zuo, Y, De, S, Steitz, T.A. | | Deposit date: | 2019-10-30 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.404 Å) | | Cite: | Structural Insights into Transcription Initiation from De Novo RNA Synthesis to Transitioning into Elongation.
Iscience, 23, 2020
|
|
6UU3
 
 | | E. coli sigma-S transcription initiation complex with a 4-nt RNA and a CTP ("Old" crystal soaked with GTP, ATP, CTP, and ddTTP for 30 minutes) | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Zuo, Y, De, S, Steitz, T.A. | | Deposit date: | 2019-10-30 | | Release date: | 2020-08-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (4.002 Å) | | Cite: | Structural Insights into Transcription Initiation from De Novo RNA Synthesis to Transitioning into Elongation.
Iscience, 23, 2020
|
|
6UU4
 
 | | E. coli sigma-S transcription initiation complex with a 3-nt RNA ("old" crystal soaked with GTP and dinucleotide GpA for 30 minutes) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zuo, Y, De, S, Steitz, T.A. | | Deposit date: | 2019-10-30 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.305 Å) | | Cite: | Structural Insights into Transcription Initiation from De Novo RNA Synthesis to Transitioning into Elongation.
Iscience, 23, 2020
|
|
6UU5
 
 | | E. coli sigma-S transcription initiation complex with a 6-nt RNA ("Old" crystal soaked with GTP, UTP, CTP, and dinucleotide GpA for 30 minutes) | | Descriptor: | DIPHOSPHATE, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Zuo, Y, De, S, Steitz, T.A. | | Deposit date: | 2019-10-30 | | Release date: | 2020-08-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (5.403 Å) | | Cite: | Structural Insights into Transcription Initiation from De Novo RNA Synthesis to Transitioning into Elongation.
Iscience, 23, 2020
|
|
6UU6
 
 | | E. coli sigma-S transcription initiation complex with a 4-nt RNA and a UTP ("Old" crystal soaked with UTP, ddCTP, and dinucleotide ApG for 30 minutes) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zuo, Y, De, S, Steitz, T.A. | | Deposit date: | 2019-10-30 | | Release date: | 2020-08-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (4.201 Å) | | Cite: | Structural Insights into Transcription Initiation from De Novo RNA Synthesis to Transitioning into Elongation.
Iscience, 23, 2020
|
|
6UU7
 
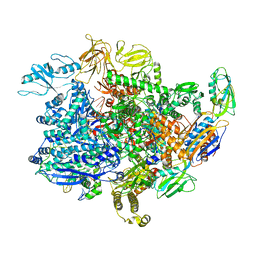 | | E. coli sigma-S transcription initiation complex with a 6-nt RNA and an NTP ("Old" crystal soaked with UTP, CTP, ddGTP, and dinucleotide ApG for 30 minutes) | | Descriptor: | 2'-3'-DIDEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Zuo, Y, De, S, Steitz, T.A. | | Deposit date: | 2019-10-30 | | Release date: | 2020-08-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Structural Insights into Transcription Initiation from De Novo RNA Synthesis to Transitioning into Elongation.
Iscience, 23, 2020
|
|
6UU8
 
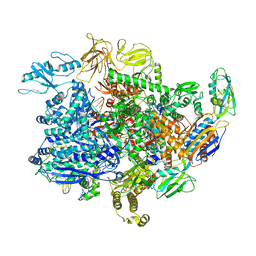 | | E. coli mutant sigma-S transcription initiation complex with a 7-nt RNA ("Fresh" mutant crystal soaked with GTP, UTP, and CTP for 30 minutes) | | Descriptor: | DIPHOSPHATE, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Zuo, Y, De, S, Steitz, T.A. | | Deposit date: | 2019-10-30 | | Release date: | 2020-08-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Structural Insights into Transcription Initiation from De Novo RNA Synthesis to Transitioning into Elongation.
Iscience, 23, 2020
|
|
6UU9
 
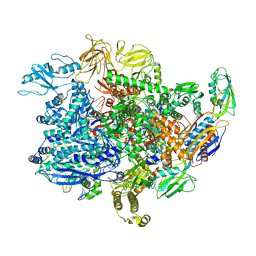 | | E. coli mutant sigma-S transcription initiation complex with an 8-nt RNA ("Fresh" mutant crystal soaked with GTP, UTP, CTP, and ddATP for 30 minutes) | | Descriptor: | 2',3'-DIDEOXYADENOSINE-5'-MONOPHOSPHATE, DIPHOSPHATE, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Zuo, Y, De, S, Steitz, T.A. | | Deposit date: | 2019-10-30 | | Release date: | 2020-08-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (5.4 Å) | | Cite: | Structural Insights into Transcription Initiation from De Novo RNA Synthesis to Transitioning into Elongation.
Iscience, 23, 2020
|
|
6UUA
 
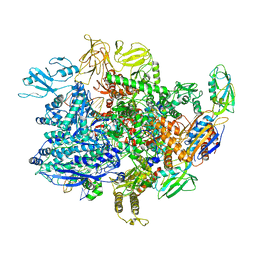 | | E. coli sigma-S transcription initiation complex with a mismatching CTP ("Fresh" crystal soaked with CTP for 2 hours) | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Zuo, Y, De, S, Steitz, T.A. | | Deposit date: | 2019-10-30 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.002 Å) | | Cite: | Structural Insights into Transcription Initiation from De Novo RNA Synthesis to Transitioning into Elongation.
Iscience, 23, 2020
|
|
6UUB
 
 | | E. coli sigma-S transcription initiation complex with a mismatching UTP ("Fresh" crystal soaked with UTP for 2 hours) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zuo, Y, De, S, Steitz, T.A. | | Deposit date: | 2019-10-30 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.955 Å) | | Cite: | Structural Insights into Transcription Initiation from De Novo RNA Synthesis to Transitioning into Elongation.
Iscience, 23, 2020
|
|
6UUC
 
 | | E. coli sigma-S transcription initiation complex with a 3-nt RNA and a mismatching ATP ("Fresh" crystal soaked with ATP for 2 hours) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Zuo, Y, De, S, Steitz, T.A. | | Deposit date: | 2019-10-30 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.096 Å) | | Cite: | Structural Insights into Transcription Initiation from De Novo RNA Synthesis to Transitioning into Elongation.
Iscience, 23, 2020
|
|
6UWI
 
 | | Crystal structure of the Clostridium difficile translocase CDTb | | Descriptor: | ADP-ribosyltransferase binding component, CALCIUM ION | | Authors: | Pozharski, E. | | Deposit date: | 2019-11-05 | | Release date: | 2020-01-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structure of the cell-binding component of theClostridium difficilebinary toxin reveals a di-heptamer macromolecular assembly.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6UZ7
 
 | | K.lactis 80S ribosome with p/PE tRNA and eIF5B | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0, ... | | Authors: | Fernandez, I.S, Huang, B.Y. | | Deposit date: | 2019-11-14 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Long-range interdomain communications in eIF5B regulate GTP hydrolysis and translation initiation.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6UZC
 
 | |







