5NRL
 
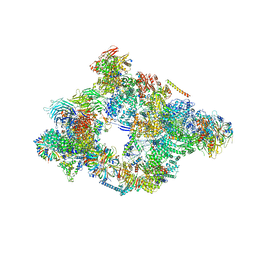 | | Structure of a pre-catalytic spliceosome. | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 23 kDa U4/U6.U5 small nuclear ribonucleoprotein component, 66 kDa U4/U6.U5 small nuclear ribonucleoprotein component, ... | | Authors: | Plaschka, C, Lin, P.-C, Nagai, K. | | Deposit date: | 2017-04-24 | | Release date: | 2017-05-31 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Structure of a pre-catalytic spliceosome.
Nature, 546, 2017
|
|
5NWY
 
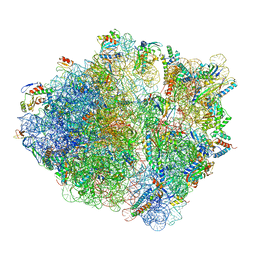 | | 2.9 A cryo-EM structure of VemP-stalled ribosome-nascent chain complex | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Su, T, Cheng, J, Sohmen, D, Hedman, R, Berninghausen, O, von Heijne, G, Wilson, D.N, Beckmann, R. | | Deposit date: | 2017-05-08 | | Release date: | 2017-07-19 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | The force-sensing peptide VemP employs extreme compaction and secondary structure formation to induce ribosomal stalling.
Elife, 6, 2017
|
|
5O09
 
 | | BtubABC mini microtubule | | Descriptor: | Bacterial kinesin light chain, GUANOSINE-5'-DIPHOSPHATE, Tubulin, ... | | Authors: | Deng, X, Bharat, T.A.M, Lowe, J. | | Deposit date: | 2017-05-16 | | Release date: | 2017-07-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Four-stranded mini microtubules formed by Prosthecobacter BtubAB show dynamic instability.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5O2R
 
 | | Cryo-EM structure of the proline-rich antimicrobial peptide Api137 bound to the terminating ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Graf, M, Berninghausen, O, Beckmann, R, Wilson, D.N. | | Deposit date: | 2017-05-22 | | Release date: | 2017-07-26 | | Last modified: | 2019-02-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | An antimicrobial peptide that inhibits translation by trapping release factors on the ribosome.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5O61
 
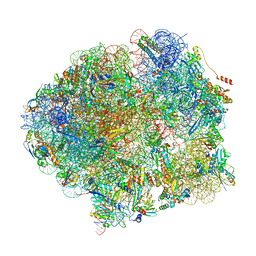 | | The complete structure of the Mycobacterium smegmatis 70S ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Hentschel, J, Burnside, C, Mignot, I, Leibundgut, M, Boehringer, D, Ban, N. | | Deposit date: | 2017-06-03 | | Release date: | 2017-07-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | The Complete Structure of the Mycobacterium smegmatis 70S Ribosome.
Cell Rep, 20, 2017
|
|
5OBM
 
 | | Crystal structure of Gentamicin bound to the yeast 80S ribosome | | Descriptor: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Prokhorova, I, Djumagulov, M, Urzhumtsev, A, Yusupov, M, Yusupova, G. | | Deposit date: | 2017-06-28 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Aminoglycoside interactions and impacts on the eukaryotic ribosome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5OJQ
 
 | | The modeled structure of of wild type extended type VI secretion system sheath/tube complex in vibrio cholerae based on cryo-EM reconstruction of the non-contractile sheath/tube complex | | Descriptor: | Haemolysin co-regulated protein, Type VI secretion protein, VipA | | Authors: | Wang, J, Brackmann, M, Castano-Diez, D, Kudryashev, M, Goldie, K, Maier, T, Stahlberg, H, Basler, M. | | Deposit date: | 2017-07-22 | | Release date: | 2017-08-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of the extended type VI secretion system sheath-tube complex.
Nat Microbiol, 2, 2017
|
|
5ON6
 
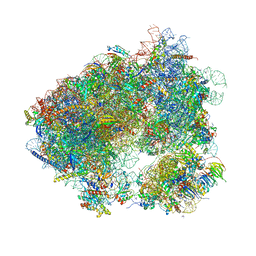 | | Crystal structure of haemanthamine bound to the 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Pellegrino, S, Meyer, M, Yusupova, G, Yusupov, M. | | Deposit date: | 2017-08-03 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.10000240787 Å) | | Cite: | The Amaryllidaceae Alkaloid Haemanthamine Binds the Eukaryotic Ribosome to Repress Cancer Cell Growth.
Structure, 26, 2018
|
|
5OQL
 
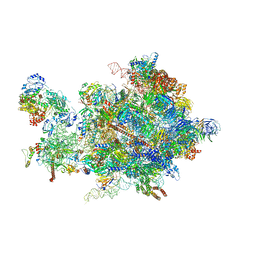 | | Cryo-EM structure of the 90S pre-ribosome from Chaetomium thermophilum | | Descriptor: | 35S rRNA, 40S ribosomal protein S1, 40S ribosomal protein S11-like protein, ... | | Authors: | Cheng, J, Kellner, N, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2017-08-12 | | Release date: | 2017-10-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | 3.2- angstrom -resolution structure of the 90S preribosome before A1 pre-rRNA cleavage.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5OT7
 
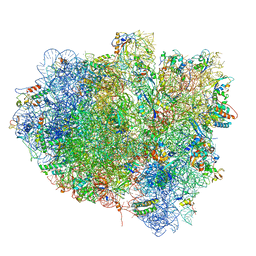 | | Elongation factor G-ribosome complex captures in the absence of inhibitors. | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Mace, K, Giudice, E, Chat, S, Gillet, R. | | Deposit date: | 2017-08-21 | | Release date: | 2018-02-14 | | Last modified: | 2018-04-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The structure of an elongation factor G-ribosome complex captured in the absence of inhibitors.
Nucleic Acids Res., 46, 2018
|
|
5SZS
 
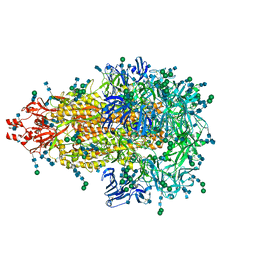 | | Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Walls, A.C, Tortorici, M.A, Frenz, B, Snijder, J, Li, W, Rey, F.A, DiMaio, F, Bosch, B.J, Veesler, D. | | Deposit date: | 2016-08-15 | | Release date: | 2016-09-14 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5T0C
 
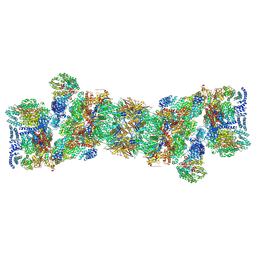 | | Structural basis for dynamic regulation of the human 26S proteasome | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Chen, S, Wu, J, Lu, Y, Ma, Y.B, Lee, B.H, Yu, Z, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2016-08-15 | | Release date: | 2016-10-19 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for dynamic regulation of the human 26S proteasome.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5T15
 
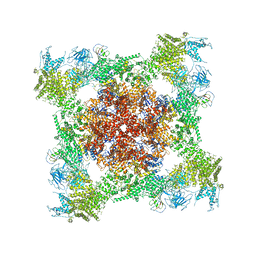 | | Structural basis for gating and activation of RyR1 (30 uM Ca2+ dataset, all particles) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1,Ryanodine receptor 1,Ryanodine receptor 1,Ryanodine receptor 1,Ryanodine receptor 1,Ryanodine receptor 1,Ryanodine receptor 1, ... | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-08-17 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5T2A
 
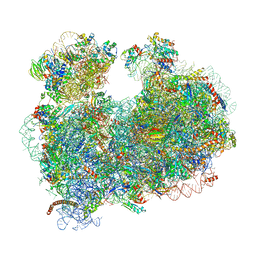 | | CryoEM structure of the Leishmania donovani 80S ribosome at 2.9 Angstrom resolution | | Descriptor: | 18S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Zhang, X, Lai, M, Zhou, Z.H. | | Deposit date: | 2016-08-23 | | Release date: | 2017-01-25 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures and stabilization of kinetoplastid-specific split rRNAs revealed by comparing leishmanial and human ribosomes.
Nat Commun, 7, 2016
|
|
5T2C
 
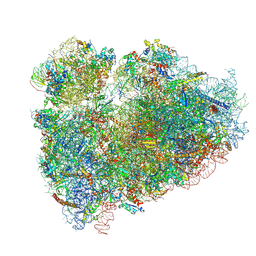 | | CryoEM structure of the human ribosome at 3.6 Angstrom resolution | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Zhang, X, Lai, M, Zhou, Z.H. | | Deposit date: | 2016-08-23 | | Release date: | 2017-01-25 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures and stabilization of kinetoplastid-specific split rRNAs revealed by comparing leishmanial and human ribosomes.
Nat Commun, 7, 2016
|
|
5T3X
 
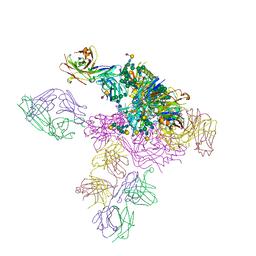 | |
5T5H
 
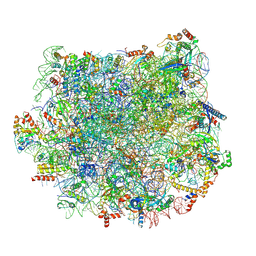 | | Structure and assembly model for the Trypanosoma cruzi 60S ribosomal subunit | | Descriptor: | 40S ribosomal protein L14, 5.8S rRNA, 5S rRNA, ... | | Authors: | Liu, Z, Gutierrez-Vargas, C, Wei, J, Grassucci, R.A, Ramesh, M, Espina, N, Sun, M, Tutuncuoglu, B, Madison-Antenucci, S, Woolford Jr, J.L, Tong, L, Frank, J. | | Deposit date: | 2016-08-31 | | Release date: | 2016-10-12 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Structure and assembly model for the Trypanosoma cruzi 60S ribosomal subunit.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5T61
 
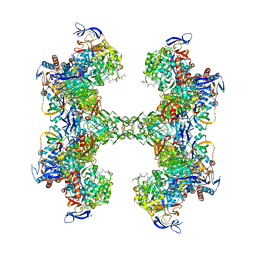 | | TUNGSTEN-CONTAINING FORMYLMETHANOFURAN DEHYDROGENASE FROM METHANOTHERMOBACTER WOLFEII, TRICLINIC FORM AT 2.55 A | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, CHLORIDE ION, HYDROSULFURIC ACID, ... | | Authors: | Wagner, T, Ermler, U, Shima, S. | | Deposit date: | 2016-09-01 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The methanogenic CO2 reducing-and-fixing enzyme is bifunctional and contains 46 [4Fe-4S] clusters.
Science, 354, 2016
|
|
5T62
 
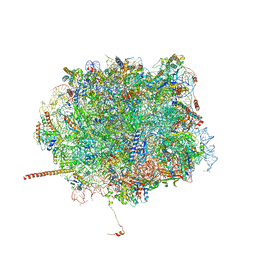 | | Nmd3 is a structural mimic of eIF5A, and activates the cpGTPase Lsg1 during 60S ribosome biogenesis: 60S-Nmd3-Tif6-Lsg1 Complex | | Descriptor: | 25S Ribosomal RNA, 5.8S Ribosomal RNA, 5S Ribosomal RNA, ... | | Authors: | Malyutin, A.G, Musalgaonkar, S, Patchett, S, Frank, J, Johnson, A.W. | | Deposit date: | 2016-09-01 | | Release date: | 2017-02-08 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Nmd3 is a structural mimic of eIF5A, and activates the cpGTPase Lsg1 during 60S ribosome biogenesis.
EMBO J., 36, 2017
|
|
5T6R
 
 | | Nmd3 is a structural mimic of eIF5A, and activates the cpGTPase Lsg1 during 60S ribosome biogenesis: 60S-Nmd3 Complex | | Descriptor: | 25S Ribosomal RNA, 5.8S Ribosomal RNA, 5S Ribosomal RNA, ... | | Authors: | Malyutin, A.G, Musalgaonkar, S, Patchett, S, Frank, J, Johnson, A.W. | | Deposit date: | 2016-09-01 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Nmd3 is a structural mimic of eIF5A, and activates the cpGTPase Lsg1 during 60S ribosome biogenesis.
EMBO J., 36, 2017
|
|
5T7V
 
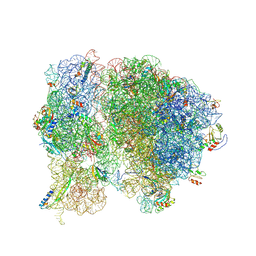 | | Methicillin Resistant, Linezolid resistant Staphylococcus aureus 70S ribosome (delta S145 uL3) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Belousoff, M.J, Lithgow, T, Eyal, Z, Yonath, A, Radjainia, M. | | Deposit date: | 2016-09-06 | | Release date: | 2017-05-31 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Basis for Linezolid Binding Site Rearrangement in theStaphylococcus aureusRibosome.
MBio, 8, 2017
|
|
5T9M
 
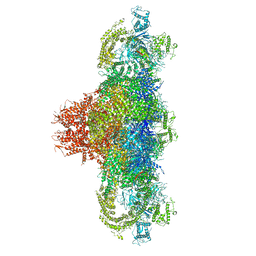 | | Structure of rabbit RyR1 (Ca2+-only dataset, class 1) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-09 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5T9N
 
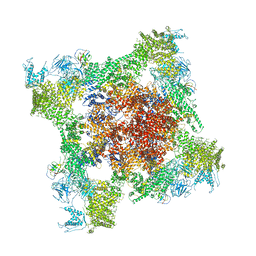 | | Structure of rabbit RyR1 (Ca2+-only dataset, class 2) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-09 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5T9R
 
 | | Structure of rabbit RyR1 (Ca2+-only dataset, class 3) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-09 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5T9S
 
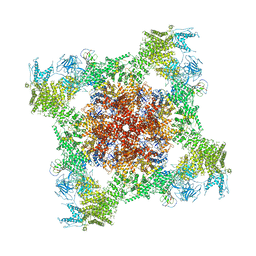 | | Structure of rabbit RyR1 (Ca2+-only dataset, class 4) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-09 | | Release date: | 2016-10-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|






